Abstract
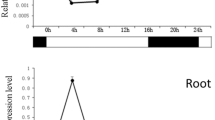
The NAC protein family is one of the novel classes of plant-specific transcription factors. In this study, two genes (BnNAC2 and BnNAC5) encoding the putative NAC transcription factors were identified in Brassica napus. Sequence analysis revealed that the deduced BnNAC proteins contain conserved N-terminal region (NAC domain) and highly divergent C-terminal domain. Yeast transactivation analysis showed that BnNAC2 could activate reporter gene expression, suggesting that BnNAC2 functions as a transcriptional activator. Quantitative RT-PCR analysis revealed that BnNAC2 was preferentially expressed in flowers, whereas BnNAC5 mRNAs accumulated at the highest level in stems. Further experimental results indicated that the two genes are high-salinity-, drought- and abscisic acid (ABA)-induced. Overexpression of BnNAC2 and BnNAC5 genes in yeast (Schizosaccharomyces pombe) remarkably inhibited the growth rate of the host cells, and enhanced the cells sensitive to high-salinity and osmotic stresses. Complementation test indicated that BnNAC5 could recover the defects such as salt-hypersensitivity and accelerated-leaf senescence of vni2 T-DNA insertion mutant. Several stress-responsive genes including COR15A and RD29A were enhanced in the complemented plants. These results suggest that BnNAC5 may perform the similar function of VNI2 in response to high-salinity stress and regulation of leaf aging.
Key message BnNAC2 and BnNAC5 are salt-, drought- and ABA-induced genes. Overexpression of BnNAC5 in Arabidopsis vni2 mutant recovered the mutant defects (salt-hypersensitivity and accelerated-leaf senescence) to the phenotype of wild type.











Similar content being viewed by others
References
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857
Aida M, Ishida T, Tasaka M (1999) Shoot apical meristem and cotyledon formation during Arabidopsis embryogenesis: interaction among the CUP-SHAPED COTYLEDON and SHOOT MERISTEMLESS genes. Development 126:1563–1570
Balazadeh S, Siddiqui H, Allu AD, Matallana-Ramirez LP, Caldana C, Mehrnia M, Zanor MI, Köhler B, Mueller-Roeber B (2010) A gene regulatory network controlled by the NAC transcription factor NAC092/AtNAC2/ORE1 during salt-promoted senescence. Plant J 62:250–264
Chen L, Ren F, Zhong H, Jiang WM, Li XB (2010) Identification and expression analysis of genes induced by high-salinity and drought stresses in Brassica napus. Acta Biochim Biophys Sin 42:154–164
Duval M, Hsieh T, Kim S, Thomas T (2002) Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Mol Biol 50:237–248
Feng Q, Zhang YJ, Hao P, Wang SY, Fu G, Huang YC et al (2002) Sequence and analysis of rice chromosome 4. Nature 420:316–320
Goff SA, Ricke D, Lan TH, Presting G et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Hao YJ, Song QX, Chen HW, Zou HF, Wei W, Kang XS, Ma B, Zhang WK, Zhang JS, Chen SY (2010) Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232:1033–1043
He XJ, Mu RL, Cao WH, Zhang ZG, Zhang JS, Chen SY (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J 44:903–916
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397
John I, Hackett R, Cooper W, Drake R, Farrell A, Grierson D (1997) Cloning and characterization of tomato leaf senescence-related cDNAs. Plant Mol Biol 33:641–651
Jung C, Seo JS, Han SW, Koo YJ, Kim CH, Song SI, Nahm BH, Choi YD, Cheong JJ (2008) Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol 146:623–635
Kim S, Kang JY, Cho DI, Park JK, Kim SY (2004) ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J 40:75–87
Kim HS, Park OB, Yoo HJ, Jung MS, Lee MS, Han JH, Kim EK, Kim HS, Lim OC, Yun D-J, Chung SW (2007) Identification of a calmodulin-binding NAC protein (CBNAC) as a transcriptional repressor in Arabidopsis. J Biol Chem 282:36292–36302
Li XB, Cai L, Cheng NH, Liu JW (2002) Molecular characterization of the cotton GhTUB1 gene that preferentially expressed in fiber. Plant Physiol 130:666–674
Li XB, Fan XP, Wang XL, Cai L, Yang WC (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17:859–875
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Shinozaki KY, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Mitsuda N, Seki M, Shinozaki K, Takagi MO (2005) The NAC transcription factors NST1 and NST2 of Arabidopsis regulate secondary wall thickenings and are required for anther dehiscence. Plant Cell 17:2993–3006
Nakashima K, Tran LSP, Nguyen DV, Fujita M, Maruyama K, Todaka D, Ito Y, Hayashi N, Shinozaki K, Shinozaki KY (2007) Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J 51:617–630
Olsen OE, Myklebust G, Engebretsen L, Holme I, Bahr R (2005) Exercises to prevent lower limb injuries in youth sports: cluster randomized controlled trial. Br Med J 330:449–452
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247
Sablowski RWM, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92:93–103
Sasaki T, Matsumoto T, Yamamoto K et al (2002) The genome sequence and structure of rice chromosome. Nature 420:312–316
Singh K, Foley RC, Onate-Sanchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436
Souer E, Houwelingen AV, Kloos D, Mol J, Koes R (1996) The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries. Cell 85:159–170
Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, Fujita M, Seki M, Shinozaki K, Shinozaki KY (2004) Isolation and functional analysis of Arabidopsis stress inducible NAC transcription factors that bind to a drought responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16:2481–2498
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Plant Biol 16:123–132
Wei W, Zhang Y, Han L, Guan Z, Chai T (2008) A novel WRKY transcriptional factor from Thlaspi caerulescens negatively regulates the osmotic stress tolerance of transgenic tobacco. Plant Cell Rep 27:795–803
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal posterior of TIR1 to promote lateral root development. Genes Dev 14:3024–3036
Yamaguchi-Shinozaki K, Shinozaki K (2006) Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu Rev Plant Biol 57:781–803
Yang SD, Seo PJ, Yoon HK, Park CM (2011) The Arabidopsis NAC transcription factor VNI2 integrates abscisic acid signals into leaf senescence via the COR/RD genes. Plant Cell 23:2155–2168
Zhang ZT, Zhou Y, Li Y, Shao SQ, Li BY, Shi HY, Li XB (2010) Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibres and involved in cell elongation. J Exp Bot 61:3331–3344
Acknowledgments
This work was supported by the project from the Ministry of Agriculture of China for transgenic research (Grant No. 2009ZX08009-117B, 2011ZX08009-003), and the Chenguang Project of Wuhan Municipality (Grant No. 200950431185).
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by Y. Lu.
H. Zhong and Q.-Q. Guo contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhong, H., Guo, QQ., Chen, L. et al. Two Brassica napus genes encoding NAC transcription factors are involved in response to high-salinity stress. Plant Cell Rep 31, 1991–2003 (2012). https://doi.org/10.1007/s00299-012-1311-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-012-1311-3