Abstract
Key message
A novel QTL (QSt.nftec-2BL) was mapped to a 0.7 cM interval on chromosome 2B. Plants carrying QSt.nftec-2BL produced higher grain yields by up to 21.4% than otherwise in salinized fields.
Abstract
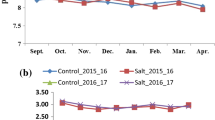
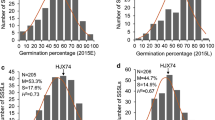
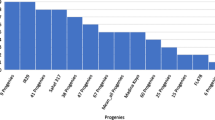
Wheat yield has been limited by soil salinity in many wheat-growing areas globally. The wheat landrace Hongmangmai (HMM) possesses salt tolerance as it produced higher grain yields than other tested wheat varieties including Early Premium (EP) under salt stresses. To detect QTL underlying this tolerance, wheat cross EP × HMM was chosen to serve as mapping population that was homozygous at Ppd (photoperiod response gene), Rht (reduced plant height gene) and Vrn (vernalization gene); thus, interference with QTL detection by these loci could be minimized. QTL mapping was conducted firstly using 102 recombinant inbred lines (RILs) that were selected from the EP × HMM population (827 RILs) for similarity in grain yield under non-saline condition. Under salt stresses, however, the 102 RILs varied significantly in grain yield. These RILs were genotyped using a 90 K SNP (single nucleotide polymorphism) array; consequently, a QTL (QSt.nftec-2BL) was detected on chromosome 2B. Then, using 827 RILs and new simple sequence repeat (SSR) markers developed according to the reference sequence IWGSC RefSeq v1.0, location of QSt.nftec-2BL was refined to a 0.7 cM (6.9 Mb) interval flanked by SSR markers 2B-557.23 and 2B-564.09. Selection for QSt.nftec-2BL was performed based on the flanking markers using two bi-parental wheat populations. Trials for validating effectiveness of the selection were conducted in salinized fields in two geographical areas and two crop seasons, demonstrating that wheat plants with the salt-tolerant allele in homozygous status at QSt.nftec-2BL produced higher grain yields by up to 21.4% than otherwise.





Similar content being viewed by others
Data availability
All data and materials described in this paper are available from the corresponding author upon request.
Code availability
Not applicable.
Abbreviations
- 2BL:
-
The long arm of chromosome 2B
- EP:
-
Early Premium, a common wheat variety
- HMM:
-
Hongmangmai, a common wheat landrace
- IWGSC:
-
International Wheat Genome Sequencing Consortium
- LOD:
-
Logarithm of the odds
- PVE:
-
Phenotypic variance explained
- QTL:
-
Quantitative trait locus/loci
- RIL:
-
Recombinant inbred line
- SNP:
-
Single nucleotide polymorphism
- SSR:
-
Simple sequence repeat
References
Asif MA, Schilling RK, Tilbrook J, Brien C, Dowling K, Rabie H, Short L, Trittermann C, Garcia A, Barrett-Lennard EG, Berger B, Mather DE, Gilliham M, Fleury D, Tester M, Roy SJ, Pearson AS (2018) Mapping of novel salt tolerance QTL in an Excalibur × Kukri doubled haploid wheat population. Theor Appl Genet 131:2179–2196
Asif MA, Garcia M, Tilbrook J, Brien C, Dowling K, Berger B, Schilling RK, Short L, Trittermann C, Gilliham M, Fleury D, Roy SJ, Pearson AS (2021) Identification of salt tolerance QTL in a wheat RIL mapping population using destructive and non-destructive phenotyping. Funct Plant Biol 48:131–140
Azadi A, Mardi M, Hervan EM, Mohammadi SA, Moradi F, Tabatabaee MT, Pirseyedi SM, Ebrahimi M, Fayaz F, Kazemi M, Ashkani S, Nakhoda B, Mohammadi-Nejad G (2015) QTL mapping of yield and yield components under normal and salt-stress conditions in bread wheat (Triticum aestivum L.). Plant Mol Biol Rep 33:102–120
Bassam BJ, Caetano-Anolles G, Gresshoff PM (1991) Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem 196:80–83
Beales J, Turner A, Griffiths S, Snape JW, Laurie DA (2007) A pseudo-response regulator is mis-expressed in photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum). Theor Appl Genet 115:721–733
Chen Y, Dai Y, Li Y, Yang J, Jiang Y, Liu G, Yu C, Zhong F, Lian B, Zhang J (2022) Overexpression of the Salix matsudana SmAP2-17 gene improves Arabidopsis salinity tolerance by enhancing the expression of SOS3 and ABI5. BMC Plant Biol 22:102
Devi R, Ram S, Rana V, Malik VK, Pande V, Singh GP (2019) QTL mapping for salt tolerance associated traits in wheat (Triticum aestivum L.). Euphytica 215:210
Díaz De León JL, Escoppinichi R, Geraldo N, Castellanos T, Mujeeb-Kazi A, Röder MS (2011) Quantitative trait loci associated with salinity tolerance in field grown bread wheat. Euphytica 181:371–383
Dubcovsky J, María GS, Epstein E, Luo MC, Dvorák J (1996) Mapping of the K+/Na+ discrimination locus Kna1 in wheat. Theor Appl Genet 92:448–454
Ellis MH, Spielmeyer W, Gale KR, Rebetzke GJ, Richards RA (2002) “Perfect” markers for the Rht-B1b and Rht-D1b dwarfing genes in wheat. Theor Appl Genet 105:1038–1042
Epstein E, Norlyn JD, Rush DW, Kingsbury RW, Kelley DB, Cunningham GA, Wrona AF (1980) Saline culture of crops: a genetic approach. Science 210:399–404
Fu D, Szücs P, Yan L, Helguera M, Skinner JS, Zitzewitz JV, Hayes PM, Dubcovsky J (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol Genet Genomics 273:54–65
Genc Y, McDonald GK, Tester M (2007) Reassessment of tissue Na+ concentration as a criterion for salinity tolerance in bread wheat. Plant Cell Environ 30:1486–1498
Genc Y, Oldach K, Gogel B, Wallwork H, McDonald GK, Smith AB (2013) Quantitative trait loci for agronomic and physiological traits for a bread wheat population grown in environments with a range of salinity levels. Mol Breed 32:39–59
Genc Y, Taylor J, Lyons G, Li Y, Cheong J, Appelbee M, Oldach K, Sutton T (2019) Bread wheat with high salinity and sodicity tolerance. Front Plant Sci 10:1280
Guerriero G, Behr M, Hausman J, Legay S (2017) Textile hemp vs. salinity: insights from a targeted gene expression analysis. Genes 8:242
Gupta PK, Balyan HS, Sharma S, Kumar R (2020) Genetics of yield, abiotic stress tolerance and biofortification in wheat (Triticum aestivum L.). Theor Appl Genet 133:1569–1602
Holland JB, Nyquist WE, Cervantes-Martinez C (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22:9–112
Ilyas N, Amjid MW, Saleem MA, Khan W, Wattoo FM, Rana RM, Maqsood RH, Zahid A, Shah GA, Anwar A, Ahmad MQ, Shaheen M, Riaz H, Ansari MJ (2020) Quantitative trait loci (QTL) mapping for physiological and biochemical attributes in a Pasban90/ Frontana recombinant inbred lines (RILs) population of wheat (Triticum aestivum) under salt stress condition. Saudi J Biol Sci 27:341–351
International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
IWGSC (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191
Jahani M, Mohammadi-Nejad G, Nakhoda B, Rieseberg LH (2019) Genetic dissection of epistatic and QTL by environment interaction effects in three bread wheat genetic backgrounds for yield related traits under saline conditions. Euphytica 215:103
Li HH, Ye GY, Wang JK (2007) A modified algorithm for the improvement of composite interval mapping. Genetics 175:361–374
Liu Y, Li D, Yan J, Wang K, Luo H, Zhang W (2019) MiR319 mediated salt tolerance by ethylene. Plant Biotechnol J 17:2370–2383
Luo Q, Zheng Q, Hu P, Liu L, Yang G, Li H, Li B, Li Z (2021) Mapping QTL for agronomic traits under two levels of salt stress in a new constructed RIL wheat population. Theor Appl Genet 134:171–189
Marone D, Russo MA, Mores A, Ficco DBM, Laidò G, Mastrangelo AM, Borrelli GM (2021) Importance of landraces in cereal breeding for stress tolerance. Plants 10:1267
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: A rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832
Mohler V, Lukman R, Ortiz-Islas S, William M, Worland AJ, van Beem J, Wenzel G (2004) Genetic and physical mapping of photoperiod insensitive gene Ppd-B1 in common wheat. Euphytica 138:33–40
Mujeeb-Kazi A, Munns R, Rasheed A, Ogbonnaya FC, Ali N, Hollington P, Dundas I, Saeed N, Wang R, Rengasamy P, Saddiq MS, De León JLD, Ashraf M, Rajaram S (2019) Breeding strategies for structuring salinity tolerance in wheat. Adv Agron 155:121–187
Mukhopadhyay P, Tyagi AK, Tyagi AK (2015) OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci Rep 5:9998
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Annu Rev Plant Biol 59:651–681
Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, Byrt CS, Hare RA, Tyerman SD, Tester M, Plett D, Gilliham M (2012) Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol 30:360–366
Narjesi V, Mardi M, Hervan EM, Azadi A, Naghavi MR, Ebrahimi M, Zali AA (2015) Analysis of quantitative trait loci (QTL) for grain yield and agronomic traits in wheat (Triticum aestivum L.) under normal and salt-stress conditions. Plant Mol Biol Rep 33:2030–2040
Newton AC, Akar T, Baresel JP, Bebeli PJ, Bettencourt E, Bladenopoulos KV, Czembor JH, Fasoula DA, Katsiotis A, Koutis K, Koutsika-Sotiriou M, Kovacs G, Larsson H, Pinheiro de Carvalho MAA, Rubiales D, Russel J, Dos Santos TMM, Vaz Patto MC (2010) Cereal landraces for sustainable agriculture. A review. Agron Sustain Dev 30:237–269
Nezhad NM, Kamali MRJ, McIntyre CL, Fakheri BA, Omidi M, Masoudi B (2019) Mapping QTLs with main and epistatic effect on Seri “M82 × Babax” wheat population under salt stress. Euphytica 215:130
Oyiga BC, Sharma RC, Baum M, Ogbonnaya FC, Léon J, Ballvora A (2018) Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat. Plant Cell Environ 41:919–935
Oyiga BC, Ogbonnaya FC, Sharma RC, Baum M, Léon J, Ballvora A (2019) Genetic and transcriptional variations in NRAMP-2 and OPAQUE1 genes are associated with salt stress response in wheat. Theor Appl Genet 132:323–346
Quamruzzaman Md, Manik SMN, Shabala S, Cao F, Zhou M (2022) Genome-wide association study reveals a genomic region on 5AL for salinity tolerance in wheat. Theor Appl Genet 135:709–721
Quarrie SA, Steed A, Calestani C, Semikhodskii A, Lebreton C, Chinoy C, Steele N, Pljevljakusic D, Waterman E, Weyen J, Schondelmaier J, Habash DZ, Farmer P, Saker L, Clarkson DT, Abugalieva A, Yessimbekova M, Turuspekov Y, Abugalieva S, Tuberosa R, Sanguineti M-C, Hollington PA, Aragues R, Royo A, Dodig D (2005) A high-density genetic map of hexaploid wheat (Triticum aestivum L.) from the cross Chinese Spring × SQ1 and its use to compare QTLs for grain yield across a range of environments. Theor Appl Genet 110:865–880
Rengasamy P (2006) World salinization with emphasis on Australia. J Exp Bot 57:1017–1023
Röder MS, Korzun V, Wandehake K, Planschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81:8014–8018
Sharma DL, Bhoite R, Reeves K, Forrest K, Smith R, Dowla MANNU (2022) Genome-wide superior alleles, haplotypes and candidate genes associated with tolerance on sodic-dispersive soils in wheat (Triticum aestivum L.). Theor Appl Genet. https://doi.org/10.1007/s00122-021-04021-8
Stam P (1993) Construction of integrated genetic linkage maps by means of a new computer package: JOINMAP. Plant J 3:739–744
Wang S, Wong D, Forrest K, Allen A, Chao S, Huang BE, Maccaferri M, Salvi S, Milner SG, Cattivelli L, Mastrangelo AM, Whan A, Stephen S, Barker G, Wieseke R, Plieske J, International Wheat Genome Sequencing Consortium, Lillemo M, Mather D, Appels R, Dolferus R, Brown-Guedira G, Korol A, Akhunova AR, Feuillet C, Salse J, Morgante M, Pozniak C, Luo MC, Dvorak J, Morell M, Dubcovsky J, Ganal M, Tuberosa R, Lawley C, Mikoulitch I, Cavanagh C, Edwards KJ, Hayden M, Akhunov E (2014) Characterization of polyploid wheat genomic diversity using a high-density 90000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wang S, Basten JC, Zeng ZB (2010) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. Accessed 24 July 2020
Xiong W, Zhao Y, Gao H, Li Y, Tang W, Ma L, Yang G, Sun J (2022) Genomic characterization and expression analysis of TCP transcription factors in Setaria italica and Setaria viridis. Plant Signal Behav 17:e2075158
Yan L, Helguera M, Kato K, Fukuyama S, Sherman J, Dubcovsky J (2004) Allelic variation at the VRN-1 promoter region in polyploid wheat. Theor Appl Genet 109:1677–1686
Yan L, Fu D, Li C, Blechl A, Tranquilli G, Bonafede M, Sanchez A, Valarik M, Yasuda S, Dubcovsky J (2006) The wheat and barley vernalization gene VRN3 is an orthologue of FT. Proc Natl Acad Sci USA 103:19581–19586
Acknowledgements
We thank Lele Luo, Dawei Wang, Xinde Lin, Qiang Ding and Jian He for providing excellent technical assistance in wheat grain yield trials. This study was supported by National Key Research and Development Program of China (2020YFD0900102), Beijing Academy of Agriculture and Forestry Sciences Sci-Tech Innovation Capacity Building Program (KJCX20200115) and National Natural Science Foundation of China (31871923; 32101702).
Author information
Authors and Affiliations
Contributions
XZ and WQ conceived the study and designed and managed the experiments. JF, WQ and XZ developed the wheat populations. XZ, XJ, WQ, JF, YZ and JR performed the phenotyping and/or SSR genotyping. XZ and XJ developed the SSR markers, constructed the genetic maps and statistically analyzed the experimental data. XZ drafted the manuscript. All authors reviewed the manuscript and provided suggestions.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical standard
The authors declare that the experiments comply with the current laws of China.
Additional information
Communicated by Peter Langridge.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Jiang, X., Zhang, Y. et al. Identification of QTL for reducing loss of grain yield under salt stress conditions in bi-parental populations derived from wheat landrace Hongmangmai. Theor Appl Genet 136, 49 (2023). https://doi.org/10.1007/s00122-023-04290-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00122-023-04290-5