Abstract
Key message
A gene encoding a laccase responsible for chartreuse onion bulb color was identified. Markers tagging this gene showed perfect linkage with bulb colors among diverse germplasm.
Abstract
To identify a casual gene for the G locus determining chartreuse bulb color in onion (Allium cepa L.), bulked segregant RNA-Seq (BSR-Seq) was performed using yellow and chartreuse individuals of a segregating population. Through single nucleotide polymorphism (SNP) and differentially expressed gene (DEG) screening processes, 163 and 143 transcripts were selected, respectively. One transcript encoding a laccase-like protein was commonly identified from SNP and DEG screening. This transcript contained four highly conserved copper-binding domains known to be signature sequences of laccases. This gene was designated AcLAC12 since it showed high homology with Arabidopsis AtLAC12. A 4-bp deletion creating a premature stop codon was identified in exon 5 of the chartreuse allele. Another mutant allele in which an intact LTR-retrotransposon was transposed in exon 5 was identified from other chartreuse breeding lines. Genotypes of molecular markers tagging AcLAC12 were perfectly matched with bulb color phenotypes in segregating populations and diverse breeding lines. All chartreuse breeding lines contained inactive alleles of DFR-A gene determining red bulb color, indicating that chartreuse color appeared when both DFR-A and AcLAC12 genes were inactivated. Linkage maps showed that AcLAC12 was positioned at the end of chromosome 7. Transcription levels of structural genes encoding enzymes in anthocyanin biosynthesis pathway were generally reduced in chartreuse bulk compared with yellow bulk. Concentrations of total quercetins were also reduced in chartreuse onion. However, significant amounts of quercetins were detected in chartreuse onion, implying that AcLAC12 might be involved in modification of quercetin derivatives in onion.






Similar content being viewed by others
Data availability
Nucleotide sequences of AcLAC12 alleles are accessible at NCBI Database under the accession numbers from OK33673 to OK336705.
References
Amawi H, Ashby CR Jr, Tiwari AK (2017) Cancer chemoprevention through dietary flavonoids: what’s limiting? Chin J Cancer 36:50
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Andersen JR, Lübberstedt T (2003) Functional markers in plants. Trends Plant Sci 11:554–560
Baek G, Kim C, Kim S (2017) Development of a molecular marker tightly linked to the C locus conferring a white bulb color in onion (Allium cepa L.) using bulked segregant analysis and RNA-seq. Mol Breed 37:94
Barbosa ICR, Rojas-Murcia N, Geldner N (2019) The Casparian strip-one ring to bring cell biology to lignification? Curr Opin Biotechnol 56:121–129
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics, btu170
Brewster JL (2008) Onions and other vegetable Alliums. CAB International, Wallingford
Cai X, Davis EJ, Ballif J, Liang M, Bushman E, Haroldsen V, Torabinejad J, Wu Y (2006) Mutant identification and characterization of the laccase gene family in Arabidopsis. J Exp Bot 57:2563–2569
Cao J, Chen W, Zhang Y, Zhang Y, Zhao X (2010) Content of selected flavonoids in 100 edible vegetables and fruits. Food Sci Technol Res 16:395–402
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Chin S, Behm CA, Mathesius U (2018) Functions of flavonoids in plant-nematode interactions. Plants 7:85
Cho Y, Kim B, Lee J, Kim S (2021) Construction of a high-resolution linkage map and chromosomal localization of the loci determining major qualitative traits in onion (Allium cepa L.). Euphytica 217:17
Clarke AE, Jones HA, Little TM (1944) Inheritance of bulb color in the onion. Genetics 29:569–575
Dixon RA, Pasinetti GM (2010) Flavonoids and isoflavonoids: From plant biology to agriculture and neuroscience. Plant Physiol 154:453–457
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
El-Shafie MW, Davis GN (1967) Inheritance of bulb color in the onion (Allium cepa L.). Hilgardia 38:607–622
Farhadi F, Khameneh B, Iranshahi M, Iranshahy M (2018) Antibacterial activity of flavonoids and their structure–activity relationship: an update review. Phytother Res 33:13–40
Fernández-Rojas B, Gutiérrez-Venegas G (2018) Flavonoids exert multiple periodontic benefits including anti-inflammatory, periodontal ligament-supporting, and alveolar bone-preserving effects. Life Sci 209:435–454
Fini A, Brunetti C, Di Ferdinando M, Ferrini F, Tattini M (2011) Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal Behav 6:709–711
Fujito S, Akyol TY, Mukae T, Wako T, Yamashita K, Tsukazaki H, Hirakawa H, Tanaka K, Mine Y, Sato S, Shigyo M (2021) Construction of a high-density linkage map and graphical representation of the arrangement of transcriptome-based unigene markers on the chromosomes of onion Allium cepa L. BMC Genom 22:481
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Window 95/98/NT. Nucl Acids Symp Ser 41:95–98
Havey (2020) Genetic mapping of chartreuse bulb color in onion. J Amer Soc Hort Sci 145:110–119
Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62:2465–2483
Holton TA, Cornish EC (1995) Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 7:1070–1083
Jaakola L (2013) New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci 18:477–483
Janusz G, Pawlik A, Świderska-Burek U, Polak J, Sulej J, Jarosz-Wilkołazka A, Paszczyński A (2020) Laccase properties, physiological functions, and evolution. Int J Mol Sci 21:966
Jo C, Kim S (2020) Transposition of a non-autonomous DNA transposon in the gene coding for a bHLH transcription factor results in a white bulb color of onions (Allium cepa L.). Theor Appl Genet 133:317–328
Khandagale K, Gawande S (2019) Genetics of bulb colour variation and flavonoids in onion. J Hort Sci Biotechnol 94:522–532
Kim S, Binzel ML, Yoo K, Park S, Pike LM (2004a) Pink (P), a new locus responsible for pink trait in onions (Allium cepa) resulting from natural mutations of anthocyanidin synthase. Mol Gen Genom 272:18–27
Kim S, Jones R, Yoo K, Pike LM (2004b) Gold color in onions (Allium cepa): a natural mutation of the chalcone isomerase gene resulting in a premature stop codon. Mol Gen Genom 272:411–419
Kim S, Jones R, Yoo K, Pike LM (2005a) The L locus, one of complementary genes required for anthocyanin production in onions (Allium cepa), encodes anthocyanidin synthase. Theor Appl Genet 111:120–127
Kim S, Yoo K, Pike LM (2005b) Development of a PCR-based marker utilizing a deletion mutation in the DFR (dihydroflavonol 4-reductase) gene responsible for the lack of anthocyanin production in yellow onions (Allium cepa). Theor Appl Genet 110:588–595
Kim S, Yoo K, Pike LM (2005c) The basic color factor, the C locus, encodes a regulatory gene controlling transcription of chalcone synthase genes in onions (Allium cepa). Euphytica 142:273–282
Kim S, Park JY, Yang T (2015) Characterization of three active transposable elements recently inserted in three independent DFR-A alleles and one high-copy DNA transposon isolated from the Pink allele of the ANS gene in onion (Allium cepa L.). Mol Genet Genom 290:1027–1037
Kim E, Kim C, Kim S (2016) Identification of two novel mutant ANS alleles responsible for inactivation of anthocyanidin synthase and failure of anthocyanin production in onion (Allium cepa L.). Euphytica 212:427–437
Kim B, Kim S (2019) Identification of a variant of CMS-T cytoplasm and development of high resolution melting markers for distinguishing cytoplasm types and genotyping a restorer-of-fertility locus in onion (Allium cepa L.). Euphytica 215:164
Kopustinskiene DM, Jakstas V, Savickas A, Bernatoniene J (2020) Flavonoids as anticancer agents. Nutrients 12:457
Kumar SV, Phale PS, Durani S, Wangikar PP (2003) Combined sequence and structure analysis of the fungal laccase family. Biotechnol Bioeng 83:386–394
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Lee YG, Cho J, Kim Y, Moon J (2016) Change in flavonoid composition and antioxidative activity during fermentation of onion (Allium cepa L.) by Leuconostoc mesenteroides with different salt concentrations. J Food Sci 81:C1385–C1393
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler Transform. Bioinformatics 25:1754–1760
Liu S, Yeh C, Tang HM, Nettleton D, Schnable PS (2012) Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS ONE 7:e36406
McCaig BC, Meagher RB, Dean JFD (2005) Gene structure and molecular analysis of the laccase-like multicopper oxidase (LMCO) gene family in Arabidopsis thaliana. Planta 221:619–636
Nickless A, Bailis JM, You Z (2017) Control of gene expression through the nonsense-mediated RNA decay pathway. Cell Biosci 7:26
Van Ooijen JW, Voorrips RE (2001) JoinMap® 3.0, Software for the calculation of genetic linkage maps. Plant Res Int, Wageningen, Netherlands
Passeri V, Koes R, Quattrocchio FM (2016) New challenges for the design of high value plant products: stabilization of anthocyanins in plant vacuoles. Front Plant Sci 7:153
Petroni K, Tonelli C (2011) Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci 181:219–229
Pourcel L, Routaboul J, Kerhoas L, Caboche M, Lepiniec L, Debeaujona I (2005) TRANSPARENT TESTA10 encodes a laccase-like enzyme involved in oxidative polymerization of flavonoids in Arabidopsis seed coat. Plant Cell 17:2966–2980
Ramsay NA, Glover BJ (2005) MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci 10:63–70
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Scarano A, Chieppa M, Santino A (2018) Looking at flavonoid biodiversity in horticultural crops: a colored mine with nutritional benefits. Plants 7:98
Seo I, Kim J, Moon J, Kim S (2020) Construction of a linkage map flanking the I locus controlling dominant white bulb color and analysis of differentially expressed genes between dominant white and red bulbs in onion (Allium cepa L.). Euphytica 216:97
Slimestad R, Fossen T, Vågen IM (2007) Onions: a source of unique dietary flavonoids. J Agric Food Chem 55:10067–10080
Song S, Kim C, Moon JS, Kim S (2014) At least nine independent natural mutations of the DFR-A gene are responsible for appearance of yellow onions (Allium cepa L.) from red progenitors. Mol Breed 33:173–186
Turlapati PV, Kim K, Davin LB, Lewis NG (2011) The laccase multigene family in Arabidopsis thaliana: towards addressing the mystery of their gene function(s). Planta 233:439–470
Van der Auwera GA, O’Connor BD (2020) Genomics in the cloud: using Docker, GATK, and WDL in Terra, 1st edn. O'Reilly Media
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, Paux E, SanMiguel P, Schulman AH (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Xu Z, Feng K, Que F, Wang F, Xiong A (2017) A MYB transcription factor, DcMYB6, is involved in regulating anthocyanin biosynthesis in purple carrot taproots. Sci Rep 7:45324
Yamazaki M, Makita Y, Springob K, Saito K (2003) Regulatory mechanisms for anthocyanin biosynthesis in chemotypes of Perilla frutescens var. crispa. Biochem Eng J 14:191–197
Yu N, Kim S (2021) Identification of Ms2, a novel locus controlling male-fertility restoration of cytoplasmic male-sterility in onion (Allium cepa L.), and development of tightly linked molecular markers. Euphytica 217:191
Zakaryan H, Arabyan E, Oo A, Zandi K (2017) Flavonoids: promising natural compounds against viral infections. Arch Virol 162:2539–2551
Zaynab M, Fatima M, Abbas S, Sharif Y, Umair M, Zafar MH, Bahadar K (2018) Role of secondary metabolites in plant defense against pathogens. Microb Pathog 124:198–202
Zhuang Y, Zuo D, Tao Y, Cai H, Li L (2020) Laccase3-based extracellular domain provides possible positional information for directing Casparian strip formation in Arabidopsis. Proc Natl Acad Sci USA 117:15400–15402
Acknowledgements
This work was supported by “Cooperative research program for agriculture science and technology development (Project No. PJ0161682021)” funded by Rural Development Administration, Republic of Korea. It was also supported by Golden Seed Project (Center for Horticultural Seed Development, No 213007-05-5-SBB10) of Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET). The authors thank Nari Yu, Ji-hwa Heo, Jeong-An Yoo, and Su-jeong Kim for their dedicated technical assistance.
Author information
Authors and Affiliations
Contributions
SeongChan Jeon performed experiments and drafted the manuscript. JiWon Han and Cheol-Woo Kim produced segregating populations. Ju-Gyeong Kim and Jae-Hak Moon performed measurements of concentrations of quercetin derivatives. Sunggil Kim organized and coordinated this research project and edited the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest relevant to this study to disclose.
Ethical standards
All experiments performed in this study were in compliance with current laws of the Republic of Korea.
Additional information
Communicated by Sandra Elaine Branham.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
122_2021_4016_MOESM1_ESM.tif
Supplementary Fig. 1 Alignment of trimmed reads produced from chartreuse and yellow bulked RNAs to sequences of AcLAC12. Images of read alignments were produced using IGV software (TIF 122 kb)
122_2021_4016_MOESM2_ESM.tif
Supplementary Fig. 2 Correlations of expression levels of all transcripts between chartreuse and yellow bulked RNAs (TIF 32 kb)
122_2021_4016_MOESM3_ESM.tif
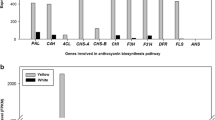
Supplementary Fig. 3 Comparison of expression levels of structural genes encoding enzymes in the flavonoid biosynthesis pathway between chartreuse and yellow bulked RNAs. Transcription levels of genes were estimated by RNA-Seq (TIF 72 kb)
122_2021_4016_MOESM4_ESM.tif
Supplementary Fig. 4 Concentrations of quercetin derivatives in yellow and chartreuse onions. A HPLC peak patterns of yellow, chartreuse, red, and white onions. HPLC analyses of red and white onions performed in a previous study (Seo et al. 2020) were included for comparison with the chartreuse onion. Standard 1: Quercetin 3,7-diglucopyranoside; Standard 2: Quercetin 3,4′-diglucopyranoside; Standard 3: Quercetin 3-glucopyranoside; Standard 4: Quercetin 4'-glucopyranoside; Standard 5: Isorhamnetin 3-glucopyranoside; and Standard 6: Quercetin aglycon. B Estimated concentrations of four quercetin derivatives in yellow, chartreuse, red, and white onions (TIF 205 kb)
Rights and permissions
About this article
Cite this article
Jeon, S., Han, J., Kim, CW. et al. Identification of a candidate gene responsible for the G locus determining chartreuse bulb color in onion (Allium cepa L.) using bulked segregant RNA-Seq. Theor Appl Genet 135, 1025–1036 (2022). https://doi.org/10.1007/s00122-021-04016-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-021-04016-5