Abstract
Key message
We identified allelic variation at two major loci, QSnb.nmbu-2A.1 and QSnb.nmbu-5A.1, showing consistent and additive effects on SNB field resistance. Validation of QSnb.nmbu-2A.1 across genetic backgrounds further highlights its usefulness for marker-assisted selection.
Abstract
Septoria nodorum blotch (SNB) is a disease of wheat (Triticum aestivum and T. durum) caused by the necrotrophic fungal pathogen Parastagonospora nodorum. SNB resistance is a typical quantitative trait, controlled by multiple quantitative trait loci (QTL) of minor effect. To achieve increased plant resistance, selection for resistance alleles and/or selection against susceptibility alleles must be undertaken. Here, we performed genetic analysis of SNB resistance using an eight-founder German Multiparent Advanced Generation Inter-Cross (MAGIC) population, termed BMWpop. Field trials and greenhouse testing were conducted over three seasons in Norway, with genetic analysis identifying ten SNB resistance QTL. Of these, two QTL were identified over two seasons: QSnb.nmbu-2A.1 on chromosome 2A and QSnb.nmbu-5A.1 on chromosome 5A. The chromosome 2A BMWpop QTL co-located with a robust SNB resistance QTL recently identified in an independent eight-founder MAGIC population constructed using varieties released in the United Kingdom (UK). The validation of this SNB resistance QTL in two independent multi-founder mapping populations, regardless of the differences in genetic background and agricultural environment, highlights the value of this locus in SNB resistance breeding. The second robust QTL identified in the BMWpop, QSnb.nmbu-5A.1, was not identified in the UK MAGIC population. Combining resistance alleles at both loci resulted in additive effects on SNB resistance. Therefore, using marker assisted selection to combine resistance alleles is a promising strategy for improving SNB resistance in wheat breeding. Indeed, the multi-locus haplotypes determined in this study provide markers for efficient tracking of these beneficial alleles in future wheat genetics and breeding activities.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat is one of the most important staple food sources worldwide, with gross production valued at around 168 billion US dollars (Food and Agriculture Organization of the United Nations 2016). However, wheat production is threatened by various bacterial, fungal and viral diseases. Parastagonospora nodorum is a devastating fungal pathogen of both bread wheat (Triticum aestivum) and durum wheat (T. durum) with disease epidemics reported in nearly all wheat producing regions with warm and humid growing conditions (Oliver et al. 2012; Francki 2013; Ficke et al. 2018). By causing damage to both wheat leaves and ears, P. nodorum can reduce yield by up to 30% (Bhathal et al. 2003). So far, regardless of resistance breeding efforts, no cultivar has shown complete resistance to P. nodorum in the field, and control of SNB still largely depends on fungicide application (Duba et al. 2018). However, intensive use of fungicides increases the risk of fungicide resistance, and the resulting reduction in available modes of action challenges the effectiveness of future chemical control (Holloman 2015). Therefore, research on host genetic resistance is needed in parallel with efforts to find new modes of action for chemical control.
P. nodorum is the model organism for necrotrophic fungal pathogens and much research has been done to characterize the interactions between P. nodorum and wheat (Oliver et al. 2012). P. nodorum can trigger plant cell death and expand infections by secreting proteinaceous necrotrophic effectors (NEs) that target dominant susceptibility loci in the wheat host (Friesen et al. 2007). This interaction between pathogen effector and host sensitivity locus is termed the ‘inverse gene-for-gene model’ (Friesen et al. 2007). Accumulation of multiple susceptibility loci in a wheat cultivar may result in higher disease severity to certain P. nodorum isolates, as some wheat susceptibilities to NEs have been shown to be quantitative and additive (Friesen et al. 2009). To date, eight P. nodorum NEs have been characterized, which interact with nine susceptibility loci distributed over seven wheat chromosomes (Ruud and Lillemo 2018). Among those, three P. nodorum NE-coding genes (ToxA, Tox1 and Tox3) (Liu et al. 2006, 2009, 2012) and two wheat susceptibility genes (Tsn1 and Snn1) (Faris et al. 2010; Shi et al. 2016) have been cloned. When used in conjunction with traditional marker-assisted selection approaches targeting SNB resistance QTL, eliminating susceptibility alleles from wheat cultivars could be a potential strategy to enhance SNB resistance breeding. For example, in Australia the ToxA-Tsn1 interaction was found to be a significant factor in field SNB susceptibility. Subsequent reduction of the ToxA sensitive wheat growing area by 13.5% between 2009 and 2013 was estimated to have saved 50 million $ in crop losses (Vleeshouwers and Oliver 2014). Genetic studies have identified numerous additional SNB resistance QTLs at the juvenile or the adult plant stages (reviewed most recently by Ruud and Lillemo 2018, with additional QTL identified in subsequent studies by Czembor et al. 2019; Francki et al. 2020; Hu et al. 2019; Lin et al. 2020a; Ruud et al. 2019), including many that have not been associated with genetic loci controlling effector sensitivity. Taken together, these map to 20 of the 21 wheat chromosomes. Therefore, phenotypic resistance in wheat is likely a result of selection against effector sensitivity alleles combined with selection for SNB resistance alleles.
Genotype by environment interaction commonly plays an important role in determining SNB resistance/susceptibility field phenotype. In addition, QTL identified using one mapping population may not necessarily be identified in another mapping population, due to differences caused by the genetic background (Langridge et al. 2001). Of the eight known NE susceptibility loci, four have been reported to co-locate with field SNB QTL: Tsn1, Snn1, Snn2 and Snn3-B1 (Friesen et al. 2009; Phan et al. 2016; Ruud et al. 2017; Ruud and Lillemo 2018). Compared to biparental populations, the genetic analysis of target traits using multi-parent advanced generation inter-cross (MAGIC) populations could be considered as more relevant for the characterization of QTL for use in breeding programs, as the multiple founders used (typically between 4 and 16) provide the possibility of capturing increased numbers of alleles at any given locus (Wei and Xu 2016; Scott et al. 2020), as well as efficiently combining founder haplotypes via multiple rounds of intercrossing. These properties of MAGIC populations allow resulting QTL to be assessed under a wider range of genetic backgrounds, and increases the chances of detecting disease resistance QTL within the framework of a single genetic mapping population (Cockram and Mackay 2018). Recently, MAGIC populations have begun to be used for numerous genetic studies of wheat disease resistance and fungal effector sensitivity (Cockram et al. 2015; Downie et al. 2018; Stadlmeier et al. 2019; Lin et al. 2020a; Corsi et al. 2020). A recent study by Lin et al. (2020a) investigated P. nodorum resistance at both the seedling and adult plant stages using a UK-relevant eight-founder wheat MAGIC resource, termed the ‘NIAB Elite MAGIC’ population (Mackay et al. 2014). Numerous QTL were identified, including QSnb.niab-2A.3, which was detected consistently across years and locations. Additionally, the observation that this QTL was identified using infiltration at the seedling stage with P. nodorum culture filtrate indicates that the adult plant SNB resistance conferred by this locus could be due to the mutation of an effector sensitivity allele (Lin et al. 2020a). The stability of QSnb.niab-2A.3 indicated that resistance alleles at this locus could be a useful target for marker assisted selection (MAS) in SNB resistance breeding.
The ‘Bavarian MAGIC winter wheat population’ (BMWpop) is an eight-founder wheat MAGIC population of German origin. Evaluating SNB disease severity in the BMWpop, which has a partially different genetic background compared to the UK ‘NIAB Elite MAGIC’, may provide additional SNB resistance loci for germplasm enhancement. In addition, if common QTL could be detected between the two MAGIC populations, MAS for such QTL could be applied with increased confidence in wider European wheat breeding programs. Lin et al. (2020a) reported that ToxA-Tsn1 and Tox3-Snn3-B1 interactions showed effects on seedling P. nodorum resistance but were not represented among QTL detected at the adult plant stage by field testing. However, Lin et al. (2020a) found that using infiltration of culture filtrate (CF) from P. nodorum isolate 203649, which possessed uncharacterized effectors, detected a QTL that was also identified for adult plant resistance in the field across years and location, QSnb.niab-2A.3 (Lin et al. 2020a). In summary, it appears that field SNB resistance is composed of interactions between known NEs and susceptibility loci, as well as additional QTLs for which the underlying mechanism controlling resistance remains unknown. The objectives of this study were to (1) identify SNB QTL in the German BMWpop MAGIC population by both seedling infiltration and field testing and compare these with QTL identified in the UK ‘NIAB Elite MAGIC’ population, (2) where phenotypic differences between the BMWpop founders for sensitivity to known P. nodorum effectors or CF are identified, phenotypically screen the population to investigate whether sensitivity QTL co-locate with adult plant SNB QTL, and (3) identify haplotypes and determine additive effects at the prioritized QTL that might help future breeding efforts to combine multiple sources of SNB resistance.
Materials and methods
Germplasm and genotypic data
The BMWpop and associated genotypic data have previously been described by Stadlmeier et al. (2018). Briefly, the population was developed at the Bavarian State Research Center for Agriculture (LfL) using eight founders (the German varieties Event, Format, BAYP4535, Potenzial, Bussard, Firl3565 and Julius, and Danish variety Ambition), selected based on multiple agronomic and disease resistance traits. The population consists of 394 F6:8 recombinant inbred lines (RILs). Together with the eight founders, the RILs were genotyped using a 15 K + 5 K Infinium iSelect single nucleotide polymorphism (SNP) array, which combined markers from the Illumina 90 K wheat SNP chip (Wang et al. 2014) and the 820 K Axiom array (Winfield et al. 2016). The resulting genotypic datasets were used by Stadlmeier and Hartl (2018) to make the BMWpop genetic map consisting of 5435 SNPs. These BMWpop resources were used here for QTL analysis of SNB resistance/sensitivity.
Field trials
Hillplot (small plots sown 50 cm apart in rows, 40 cm between rows) trials were conducted for SNB leaf blotch over three seasons (2016, 2017, 2018) at Vollebekk research station in Ås, Norway. The germplasm used for each trial consisted of the BMWpop (394 RILs and eight founders) as well as three control varieties: Jenga (relatively high resistance), Arina (moderately resistant) and Tarso (susceptible). An augmented trial design was used where the eight founders and the three controls were laid out using an alpha lattice with eight replicates, giving two checks in each row of twelve plots. Then the RILs, mostly unreplicated, were distributed randomly in the remaining field plots. Field trials were sown in autumn, established over the winter, and phenotyped the following summer as they progressed to maturity. Naturally P. nodorum-infected straw harvested from the most susceptible lines in the previous field season were used as inoculum and was applied to the field trials before stem elongation in spring. Mist irrigation was applied for 5 min every half hour from 10 am to 8 pm to enhance infection. Mist irrigation started at the same time as the inoculum was applied to the field and ended after the final scoring had been done. The selective fungicide Forbel 750 (Bayer Crop Science, a.i.: phenpropimorph) was applied to the field trials every three weeks at the full recommended dose rate to control infections of stripe rust and powdery mildew. Forbel 750 has little to no effect on P. nodorum infection.
Phenotypic evaluation of SNB leaf blotch severity in the field
SNB leaf blotch severity was scored via visual estimation of the percentage of diseased leaf area in each hillplot canopy. In 2016 and 2017, leaf blotch severity was assessed three times. The first disease scoring was carried out when the diseased area of the canopy reached around 50% for the most susceptible lines/controls, followed by approximately weekly assessments. Due to hot and dry weather, plant development was strongly accelerated in 2018, resulting in disease scoring being undertaken only twice. The first scoring followed the same criteria as described above, while the second scoring was done when the most susceptible lines reached 100% disease severity. Plant height (PH) and days to heading (DH) were also assessed each year. PH (cm) was measured from ground to bottom of the wheat ears, and DH was scored when most plants within a hillplot had ears fully emerged (Zadoks` growth stage 55, GS55).
Culture filtrate and Tox3 preparation
The P. nodorum isolate 203649 used for culture filtrate infiltration of the BMWpop was the same as described by Lin et al. (2020a). This isolate was selected to screen the BMWpop as it is a local Norwegian isolate relevant to our field trial locations in Norway. Furthermore, this isolate does not produce any of the three cloned effectors (ToxA, Tox1 and Tox3), thus potentially helping the identification of novel effector sensitivity loci beyond Tsn1, Snn1 and Snn3-B1. Following the methods described by Friesen and Faris (2012), the isolate was grown in liquid Fries 3 medium and after three-week stationary growth, the culture filtrates were filter-sterilized. Previous studies showed that sensitivities to ToxA and Tox3 were positively correlated with SNB susceptibility in the field in Norway (Ruud et al. 2017, 2018). However, all eight founders of BMWpop are insensitive to the ToxA effector. Therefore, infiltration experiments for ToxA were not carried out in this study. For Tox3 effector production, Tox3 was expressed in Pichia pastoris as described by Tan et al. (2014). The semi-purified Tox3 effector was desalted in 20 mM sodium phosphate (pH 7.0) and freeze-dried for storage. Prior to use, ultra-pure water was used for re-suspension of Tox3.
Seedling infiltration
Three to four seeds of each of the BMWpop RILs and the eight founders were sown in cones (Stuewe and sons, Tangent, Orlando, USA) filled with peat soil (Gartnerjord, Tjerbo, Norway). Seedlings were grown in a greenhouse with 16 h light per day, temperature 20/16 °C (day/night) and 65% relative humidity for 14 days. Approximately 50 μL of the culture filtrate or Tox3 effector was infiltrated into the second leaf of two-week-old seedlings using a 1-mL syringe with needle removed. Reactions to culture filtrate or Tox3 effector were scored seven days after infiltration using a 0–3 scale (Friesen and Faris 2012) where 0 represents completely insensitive, 1 represents mottled chlorosis, 2 represents complete chlorosis, and 3 represents necrosis. The experiment was conducted with three biological replicates of each RIL and six to nine biological replicates for each of the eight founders.
Statistical analysis
The PROC MIXED procedure in SAS v.9.4 (SAS Institute Inc.) was used to calculate mean disease severity, PH and DH of each genotype within each year. For the analysis of the field trials within each year, multi-linear regression with PH and DH as covariates was carried out in RStudio version 1.1.442 (RStudio Team 2015) to determine whether PH and/or DH affected leaf blotch disease severity with the model:
The corrected disease severities were obtained by subtracting the residuals from the linear regression model with PH and/or DH as covariate, when PH and/or DH was significantly (p < 0.0001) correlated with leaf blotch disease severity using “resid” function by R. Shapiro–Wilk tests were carried out in RStudio to test for normality of mean/corrected disease severity.
QTL analysis
A subset of 2804 SNP markers previously assigned to unique map positions in the BMWpop genetic map (Stadlmeier et al. 2019) were used for interval mapping (IM) and composite interval mapping (CIM). Founder probabilities were calculated using the function ‘mpprob’ in the R/mpMap package V2.0.2 (Huang and George 2011) at the threshold of 0.7. IM was carried out using the function ‘mpIM’ in R/mpMap with the founder haplotype probabilities obtained from the previous step. CIM was undertaken using either 5 (CIM-cov5) or 10 (CIM-cov10) cofactors. 1000 simulations of the phenotypic dataset were conducted and used to obtain an empirical distribution of genome-wide significance p values based on a null QTL hypothesis. The significance threshold was then determined by the genome-wide significance p value at the threshold level α < 0.05, on a trait by trait basis. All detected QTL were then fitted in a full model using the function ‘fit’ to obtain additive founder effects (relative to the founder, Julius) and the phenotypic variation (R2) explained by each QTL. The supporting interval of each QTL was defined as markers with −log10(p) value ± 0.5 of the peak marker’s −log10(p) value. CIM-cov5 and CIM-cov10 were carried out to further confirm and refine the genetic map locations of the QTL detected by IM. In addition, QTL mapping via identity-by-descent (IBD) analysis was undertaken to support the outcome of IM using all 5435 mapped SNP markers, based on a regression model against the founder haplotype probabilities of each marker. The founder haplotype probabilities were calculated as described above, and the additive founder effects were estimated relative to the founder, Julius. R/q value package was used to correct for multiple testing of IBD with a significant threshold q = 0.05. Flanking DNA sequences for SNP markers were obtained from websites https://triticeaetoolbox.org and https://www.cerealsdb.uk.net. Physical map positions of SNP markers in the cv. Chinese Spring wheat reference genome assembly, RefSeq v1.0 (International Wheat Genome Sequencing Consortium (IWGSC) et al. 2018), were obtained via BLASTn analysis using the website https://urgi.versailles.inra.fr/blast_iwgsc/?dbgroup=wheat_iwgsc_refseq_v1_chromosomes&program=blastn. Genetic linkage groups overlaid with the positions of QTL intervals were graphically displayed using Mapchart 2.32 (Voorrips 2002).
Haplotype analysis for QTL QSnb.nmbu-2A.1 and QSnb.nmbu-5A.1
Five markers within the QSnb.nmbu-2A.1 QTL interval from 2018 (QSnb.nmbu-2A.1/2018) and with the highest −log10(p) values were selected to construct haplotypes. Six most significant markers located at the overlapping supporting interval of QSnb.nmbu-5A.1 detected in both 2016 and 2018 were selected to construct haplotypes. As the haplotype effect of QSnb.nmbu-5A.1 was contributed mainly by allelic differences at marker wsnp_Ex_c898_1738424, for this QTL the comparison was based on the allele effect of marker wsnp_Ex_c898_1738424 alone. The mean disease severities from 2016 and corrected disease severities from 2017 and 2018 for each haplotype in the BMWpop RILs were calculated and compared by Kruskalmc test (p < 0.05) using the R/pgirmess package (Giraudoux 2018). An additional haplotype analysis of QTL QSnb.nmbu-2A.1/2016 was conducted using the peak marker BobWhite_c3833_815 and the closely linked marker AX-94825088 for validating the founder effect of the founder, Event.
Analysis of additive effects
Genotypes possessing either only the susceptible haplotype 4 at QSnb.nmbu-2A.1/2018 or only the susceptible allele for marker wsnp_Ex_c898_1738424 at QSnb.nmbu-5A.1, were grouped together as carrying one resistant allele. Genotypes that carried both susceptible alleles were grouped as carrying zero resistant alleles, while the remaining genotypes were grouped as carrying two resistant alleles. Comparison of disease severities between genotype groups was conducted using the same method as described for haplotype analysis.
Results
Phenotypic evaluation of SNB field resistance
The three varieties used as SNB controls performed as expected, with Tarso, Arina and Jenga showing high, medium and low SNB infection in all trials, respectively (Table S1). Among the eight BMWpop founders, Bussard and Event showed the highest mean leaf blotch disease severity (Fig. 1a). Broad and transgressive variation for leaf blotch severity was observed among BMWpop RILs (Fig. 1b). PH was not significantly correlated with leaf blotch severity in any of the three years studied, whereas DH was significantly correlated with disease severity in both 2017 (r = −0.31, p < 0.0001) and 2018 (r = −0.24, p < 0.0001) (Table 1). The mean leaf blotch severities were all significantly (p < 0.0001) correlated between years, with phenotypic correlation coefficients ranging from 0.36 (2017–2018) to 0.47 (2016–2017) (Table 1). As neither PH nor DH was significantly correlated with disease severity in 2016, the mean disease severities from 2016 were used directly for both QTL and haplotype analysis, while disease severity data from both 2017 and 2018 were corrected for DH effects. The mean disease severity data were not normally distributed (p < 0.0001), being skewed toward lower disease severity in all three years (Fig. 1b).
Phenotypic evaluation of seedling infiltration
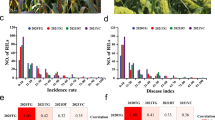
Founder reactions to infiltration with culture filtrate of P. nodorum isolate 203649 are shown in Fig. 2a. Ambition, BAYP4535 and Event were completely insensitive (score 0). Two of the nine replicates tested for Julius showed a weak sensitive reaction (score 1), the remaining seven were completely insensitive (score 0), whereas Bussard, Firl3565, Format and Potenzial showed higher sensitivity (scores between 1 and 2). However, transgressive segregation was observed in the population (Fig. 2b): 8.4% of the RILs showed a culture filtrate sensitivity score > 2, which exceeded the sensitivity range of all eight founders (Fig. 2a). 59.4% of the RILs were insensitive (score < 1), while 32% of RILs showed intermediate reactions (1 < score < 2) (Fig. 2b).
a Reactions of BMWpop founders to infiltration with culture filtrates of P. nodorum isolate 203649 (left) and Tox3 effector (right). Error bars indicate the standard deviation of mean reaction type for each parent. 9 replicates were used for infiltration with isolate 203649, while 6 replicates were used for infiltration with Tox3. b Histogram of the reactions of BMWpop RILs to infiltration with culture filtrates of P. nodorum isolate 203649 (left) and Tox3 effector (right)
Infiltration with the P. nodorum effector Tox3 identified three founders (Bussard, Julius and Potenzial) to be highly sensitive (score 3), whereas Ambition, Event and Firl3565 were completely insensitive (score 0) (Fig. 2a). Two founders showed intermediate reactions, with Format being relatively sensitive (score 2) and BAYP4535 being relatively insensitive with a mean score < 1 (Fig. 2a). Sixty-four percent of the RILs showed strong sensitivity to Tox3 (score 3), whereas 18% of the RILs showed complete insensitivity (Fig. 2b). The remaining 18% of the RILs showed intermediate sensitivity (Fig. 2b).
Genetic analysis of SNB resistance/sensitivity
Four significant QTL were detected for field leaf blotch resistance/sensitivity on chromosomes 2A, 2B, 2D and 5A, each explaining between 5 and 7% of the phenotypic variation (Table 2). In this study, two QTL were initially detected for leaf blotch resistance/susceptibility on chromosome 2A: QSnb.nmbu-2A.1/2018 and QSnb.nmbu-2A.1/2016 (Fig. 3a). QSnb.nmbu-2A.1/2018 was the most significant of the two (−log10(p) = 3.4) and explained 5.4% of the phenotypic variation (peak marker wsnp_CAP8_c2677_1394934, located at 146.2 cM on the genetic map and 603.5 Mb on the physical map) (Table 2 and Fig. 3a, b). At this QTL, the Event allele increased the corrected disease severity relative to Julius by 5.4%, whereas alleles from all but one of the remaining founders decreased the corrected disease severity by > 3% relative to Julius—notably Format and Bussard, with disease reductions of 6.2% and 4.7%, respectively (Fig. 3c). QSnb.nmbu-2A.1/2016 (–log10(p) = 2.8) was detected in the 2016 trial on chromosome 2A at 190 cM (692 Mb) and explained 5.2% of the phenotypic variation (Table 2 and Fig. 3a), with the allele from Event also contributing the most to susceptibility (increasing the mean disease severity relative to Julius by 9.01%). However, in contrast to QSnb.nmbu-2A.1/2018, the Format allele had a relatively high increasing effect on mean disease severity (Fig. 3c). QSnb.nmbu-2A.1/2018 and QSnb.nmbu-2A.1/2016 were firstly considered as two distinct QTL since their respective QTL peaks were located approximately 40 cM apart on BMWpop genetic map. However, on the wheat physical map, the QSnb.nmbu-2A.1/2016 interval was located within that of QSnb.nmbu-2A.1/2018 (Fig. 3b). Additionally, the predicted founder effect of these two QTL was similar, where Event contributed the most to disease susceptibility (Fig. 3c). As these QTL are close to the highly non-recombining region (Table 3, Fig. 3b), the balance of evidence is not sufficient to state that these two QTL are different. We therefore subsequently treat them as a single QTL here. However, further validation is required to confirm this assumption.
Summary information for leaf blotch QTL identified for leaf blotch on chromosome 2A in the BMWpop population. a Results of QTL scan using interval mapping (IM). Peak marker QTL detected in 2018 is indicated in pink, while QTL detected in 2016 are indicated in green. b Plot of the genetic (Stadlmeier et al. 2018) versus physical map (IWGSC RefSeq v1.0) position of SNPs mapped to chromosome 2A in the BMWpop. Markers within the support interval of QSnb.nmbu-2A.1/2016 and QSnb.nmbu-2A.1/2018 are indicated in green and blue, respectively. c Predicted founder effects for QSnb.nmbu-2A.1/2016 and QSnb.nmbu-2A.1/2018, relative to the founder Julius. Error bars indicate the standard error of the estimated founder effects
Another robust SNB resistance/sensitivity QTL identified in the BMWpop, QSnb.nmbu-5A.1 on the long arm of chromosome 5A, was detected in both 2016 and 2018 and explained 6.7% and 5.0% of the phenotypic variation, respectively (Table 2, Fig. 4). The founder effects of QSnb.nmbu-5A.1 were not conclusive for the 2 years (data not shown). QSnb.nmbu-2B.1 (Fig. 4) on the long arm of chromosome 2B (−log10(p) = 3.5, R2 = 6.9%) and QSnb.nmbu-2D.1 (Fig. 4) on the short arm of chromosome 2D (−log10(p) = 2.6, R2 = 7.0%) were also significant, but only detected in single years (Table 2).
SNB QTL identified on chromosomes 2B, 2D, 5A, 5B, 6A and 7B. Results of IM are shown. QTL and permutated thresholds (−log10(p)) are colored by trait; field season 2016: black, field season 2017: blue, field season 2018: red, Culture filtrate infiltration of isolate 203649: green, Infiltration of Tox3: brown. Peak markers are indicated in pink
Genetic analysis of seedling infiltration
Six significant QTL were detected for greenhouse infiltration experiments, on chromosomes 5A, 5B, 6A and 7B. Two QTL were identified for Tox3 infiltration and four via infiltration using culture filtrate from P. nodorum isolate 203649. For Tox3 infiltration, QTox3.nmbu-5B.1 co-located with the major Snn3-B1 Tox3 sensitivity locus on the short arm of chromosome 5B (p = 0, R2 = 36%, peak marker wsnp_JD_rep_c48937_33188230, located at 3.05 cM/ 14.5 Mb) (Table 2, Fig. 4). In addition, another Tox3 sensitivity QTL was detected on the long arm of chromosome 6A (QTox3.nmbu-6A.1, −log10(p) = 4, R2 = 5.5%) (Table 2). However, QTox3.nmbu-6A.1 was detected using IM and IBD only, and not via CIM-cov5 or -cov10. The most significant QTL for sensitivity to culture filtrate infiltration with isolate 203649 was located on chromosome 7B (QInf.nmbu-7B.1: −log10(p) = 14.9, peak marker wsnp_Ex_c56425_58548596 at 176.9 cM/683.5 Mb) and accounted for 17.1% of the phenotypic variation (Table 2, Fig. 4). Three additional QTL less significant than QInf.nmbu-7B.1 were also detected for sensitivity to culture filtrate infiltration on chromosomes 5A, 5B and 6A (Table 2).
Haplotype analysis and additive effects of SNB resistance QTL QSnb.nmbu-2A.1 and QSnb.nmbu-5A.1
Markers used for haplotype construction at QSnb.nmbu-2A.1/2018 are listed in Table 3. In total, the five SNPs used defined five haplotypes. Consistent significant difference (p < 0.05) of mean/corrected disease severity was observed between not only haplotypes 3 and 4, but also between haplotypes 4 and 5 in all tested years (Fig. 5). Haplotype 4 was also always the most susceptible haplotype, with approximately 11% higher disease severity compared to haplotype 3 and haplotype 5 (Table S2). Haplotype 4 originated from the founder Event. While relatively resistant haplotype 3 originated from BAYP4535 and Firl3565, and haplotype 5 originated from Bussard and Format. The same haplotype analysis was undertaken using the phenotypic data from culture filtrate infiltration with isolate 203649, however, no significant difference of disease severity between the QSnb.nmbu-2A.1/2018 haplotypes was observed (data not shown). Additional haplotype analysis of QSnb.nmbu-2A.1/2016 using two significant markers defined three haplotypes, where the haplotype two originating from the founder Event always showed higher susceptibility. Significant differences between haplotype 1 and 2 (p < 0.05) were observed in two out of the three years tested (Fig. S1). For QTL QSnb.nmbu-5A.1, the allele effect of marker wsnp_Ex_c898_1738424 (210.95 cM, 574 Mb) on SNB disease severity was significant (p < 0.05) in all tested years (Fig. 6). The susceptible allele was inherited from Format, while the remaining founders carry the resistance allele (Fig. 6). Figure 7 shows the decrease in SNB disease severities by stacking resistant alleles. In all tested years, significant differences in mean/corrected SNB disease severity were observed between genotypes carrying no resistant allele and those carrying 1 or 2 resistant alleles (Fig. 7). In addition, significant additive effect of stacking resistant alleles was observed in 2016 (Fig. 7a).
Haplotype analysis for BMWpop leaf blotch QTL QSnb.nmbu-2A.1/2018. a Haplotype analysis of mean disease severity in field season 2016. b, c Haplotype analysis of corrected disease severity in field season 2017 and 2018, respectively, and the mean disease ratings for the 8 founders are indicated. Haplotypes labeled with same letter represented no significant differences between haplotype disease severities as detected by Kruskalmc test (p < 0.05). d Genotype of each haplotype based on five SNP markers. SNP marker names are listed in order as below: wsnp_CAP8_c2677_1394934, AX-95661975, RAC875_c38018_278, AX-94508462, BS00090569_51
Allele effect analysis of marker wsnp_Ex_c898_1738424 for BMWpop SNB QTL QSnb.nmbu-5A.1. a Allele effect of mean disease severity in field season 2016. b, c Allele effect of corrected disease severity in field season 2017 and 2018, respectively, and the mean disease ratings for the eight founders are indicated. Genotypes labeled with same letter represented no significant differences between haplotype disease severities as detected by Kruskalmc test (p < 0.05)
Analysis of additive effects for QTL QSnb.nmbu-2A.1/2018 and QSnb.nmbu-5A.1. a Additive effect analysis of mean disease severity in field season 2016. b, c Additive effect analysis of corrected disease severity in field seasons 2017 and 2018, respectively. Genotypes labeled with same letter represented no significant differences between haplotype disease severities as detected by Kruskalmc test (p < 0.05)
Discussion
In this study, SNB field trials using a German MAGIC population (‘BMWpop’) were carried out over three seasons from 2016 to 2018 at Vollebekk field station in Ås, Norway, side by side with the trials previously reported for the UK MAGIC population (‘NIAB Elite MAGIC’) (Lin et al. 2020a). The two MAGIC populations were subjected to the same P. nodorum field population and similar environmental influences. However, QTL identified in the ‘NIAB Elite MAGIC’ population may not necessarily be detected in the BMWpop, despite the similar field environments the trials were conducted under. Except for cv. Ambition which originated from a Danish breeding program (Nordic Seed), all BMWpop founders were commercially released or bred for release in Germany (Stadlmeier et al. 2018), while founders of the ‘NIAB Elite MAGIC’ were all released in the UK (Mackay et al. 2014). In addition, the two MAGIC populations were genotyped using different SNP arrays, with 1335 SNP markers in common for direct comparison of genetic maps. Thus, in addition to the use of different founders between the two populations, differences in marker density, RIL numbers and crossing designs may also result in differences in power and precision with which to detect QTL. Despite all these factors, two adult plant SNB resistance/sensitivity QTL were detected in common between the two populations: QSnb.nmbu-2A.1 and QSnb.nmbu-2D.1.
The BMWpop 2A QTL QSnb.nmbu-2A.1 was identified as a robust QTL for SNB leaf blotch susceptibility in the UK MAGIC population across multiple years (QSnb.niab-2A.3) (Lin et al. 2020a). The QSnb.nmbu-2A.1/2018 interval overlapped with that of QSnb.niab-2A.3, and the peak marker of QSnb.nmbu-2A.1/2018 was just ~ 2 Mb away from that of QSnb.niab-2A.3 for 2016 in Norway (Table 3). Haplotype analysis of QSnb.niab-2A.3 has previously confirmed the robustness of this QTL across years and locations in the ‘NIAB Elite MAGIC’ population (Lin et al. 2020a). Interestingly, haplotype analysis also confirmed the consistent effect of BMWpop QTL QSnb.nmbu-2A.1/2018 for leaf blotch susceptibility in all three tested years and QSnb.nmbu-2A.1/2016 for two years (Fig. 5, Fig. S1). When comparing genetic maps of the two MAGIC populations, two common markers BS00090569_51 and RAC875_c38018_278 were found within the supporting intervals of these chromosome 2A QTL in both the UK MAGIC (QSnb.niab-2A.3) and German MAGIC (QSnb.nmbu-2A.1/2018) populations. Both markers were among the most significant markers detected for QSnb.nmbu-2A.1/2018 and were used for constructing QSnb.nmbu-2A.1/2018 haplotypes. QSnb.nmbu-2A.1/2018 haplotype 4 and QSnb.nmbu-2A.1/2016 haplotype 2, which were inherited from the founder Event, showed significantly higher disease severity (Fig. 5, Fig. S1). This observation also fitted the predicted founder effects at this QTL, with Event contributing the most to leaf blotch susceptibility (Fig. 3c). Therefore, we hypothesize that the susceptible haplotype from founders Xi19 and Rialto in the UK MAGIC population and the susceptible haplotype from Event in the German MAGIC population may carry the same susceptibility allele. However, while pedigree analysis shows that Xi19 was a result of a cross between the varieties Rialto and Cadenza, no pedigree information for Event could be identified to confirm possible common allelic origin within established wheat pedigree resources (e.g., Fradgley et al. 2019). Moreover, one flanking marker for the P. nodorum seedling resistance/sensitivity QTL QSnb.cur-2AS.1 reported by Phan et al. (2016) also aligned to the interval defined by QSnb.nmbu-2A.1 (Table 3). However, since QSnb.cur-2AS.1 encompasses a large physical interval (from 112 to 709 Mb), further meaningful comparison is not possible.
QSnb.nmbu-5A.1 located between 558.693–571.683 Mb on chromosome 5A was the second BMWpop QTL to be significant in more than one year. In addition, the allelic effect using marker wsnp_Ex_c898_1738424 was significant for all tested years in our study, where only the founder Format carries the susceptibility allele (Fig. 6). Given the discriminatory nature of this SNP in our eight founders, these results highlight the potential of this marker for application in marker-assisted selection. Two adult plant SNB resistance/susceptibility QTL have previously been reported on chromosome 5A (Friesen et al. 2009; Francki et al. 2020) (Table S3). Of these, our SNB QTL QSnb.nmbu-5A.1 (558.7–571.7 Mb) overlaps with the location of a locus conferring resistance/susceptibility to SNB in the bi-parental population BR34 × Grandin (~ 558.340 Mb) (Friesen et al. 2009). The BMWpop QTL QSnb.nmbu-2D.1 (14.6–15.1 Mb) detected in 2016 co-located with the ‘NIAB Elite MAGIC’ QTL QSnb.niab-2D.1 (14.8–27.8 Mb), reported by Lin et al. (2020a) to be located near the well characterized Tox2 sensitivity locus Snn2 (6.2–12.4 Mb) (Zhang et al. 2009) and QSnb.cur-2DS (14.3–37.0 Mb) (Phan et al. 2016) (Table S4).
QTL QSnb.niab-2A.3 was recently identified by culture filtrate infiltration with isolate 203649 in the ‘NIAB Elite MAGIC’ population, with the same haplotype effect observed for both field resistance and sensitivity to infiltration and inoculation with isolate 203649 (Lin et al. 2020a). This is the same isolate as used here for CF infiltration of the BMWpop. However, no CF QTL on chromosome 2A were identified in the BMWpop. Rather, CF infiltration identified QTL on chromosomes 5A, 5B and 7B. The reason why CF with isolate 203649 did not identify the 2A QTL in the BMWpop even though an SNB leaf blotch QTL was detected at this locus is not clear, and could be due to various reasons such as interactions between the CF effector complement and the genetic background of the BMWpop, or differences in the expression of effectors within different batches of CF. Interestingly, the ʻweakʼ ‘NIAB Elite MAGIC’ SNB resistance QTL QSnb.niab-7B.2 (−log10(p) = 2.91, R2 = 5.83%) was detected as a major BMWpop QTL for culture filtrate infiltration (QInf.nmbu-7B.1: −log10(p) = 14.9, R2 = 17.1%). The QTL interval of QInf.nmbu-7B.1 is located within that of QSnb.niab-7B.2 on the physical map, and their peak markers were located ~ 4 Mb apart (Table S5). Phan et al. (2016) found that Tox3 expression levels were increased when the Tox1 gene was knocked out in P. nodorum isolate SN15, indicating that Tox3 expression was suppressed by Tox1. It is possible that the expression of the uncharacterized P. nodorum effector which interacted with the QInf.nmbu-7B.1 locus in the BMWpop may have suppressed the expression of the uncharacterized effector which has been previously shown via culture filtrate infiltration in the UK MAGIC population to interact with QSnb.niab-2A.3 (Lin et al. 2020a). Recently, Peters Haugrud et al. (2019) also reported that effects caused by the NE-host inverse gene for gene interactions varied from epistatic to additive and depended on the genetic backgrounds of both host and pathogen. Therefore, the different genetic background of the host populations might result in the phenomenon where QSnb.niab-2A.3 was detected in ‘NIAB Elite MAGIC’ via culture filtrate infiltration but not in the BMWpop, despite infiltration with culture filtrate using the same isolate. In addition, the expression level of the uncharacterized NE which interacted with QSnb.niab-2A.3 in ‘NIAB Elite MAGIC’ might be low, and thus the detection of this interaction could be masked by the interaction of QInf.nmbu-7B.1 and the other uncharacterized NE. Similar to CF infiltration results, even though BMWpop segregates for the Snn3-B1 locus and the Tox3 gene is common in the Norwegian P. nodorum population (Ruud et al. 2018), the Snn3-B1 QTL was not detected in our field testing. This observation could be explained by a hypothesis proposed by Peters Haugrud et al. (2019) where P. nodorum isolates might not express all of the NE genes they harbor. Alternatively, the pathogen population occurring in the field lacked Tox3 or an epistatic effect may exist between unknown NE-Snn and Tox3-Snn3-B1 interactions under the field condition. Clearly the situation is relatively complex, and further studies are required to disentangle P. nodorum effector–wheat susceptibility interactions as well as P. nodorum effector–effector interactions in order to determine which SNB QTL are dependent on host-NE interactions.
In order to identify additional host genetic loci controlling sensitivity to new P. nodorum effectors, and to compare the locations of these to SNB QTL, we screened the BMWpop for sensitivity to CF from a local Norwegian P. nodorum isolate that lacked the three cloned effectors (ToxA, Tox1 and Tox3). None of the CF sensitivity QTL co-located with SNB leaf blotch QTL in this population. This could be because there were no effectors present in the CF, or because the BMWpop founders do not contrast for sensitivity to the hypothesized effectors. Alternatively, the isolate we used may not be representative of those responsible for the natural field infection conditions under which we investigated SNB resistance. Indeed, as our recent research finds the Norwegian P. nodorum population to be genetically diverse (lacking in obvious genetic populations substructure, and collectively containing all eight possible combinations for the presence/absence of the three cloned effector genes) (Lin et al. 2020b), it is likely that this latter hypothesis is correct. Future work investigating possible correlation between NE-Snn interactions and SNB in the BMWpop would therefore benefit from CF assessment using isolates obtained from the naturally infected experimental trials under study. In addition to screening for CF sensitivity, we also phenotyped the BMWpop for sensitivity to Tox3. This identified the major sensitivity locus Snn3-B1, as well as a minor sensitivity QTL on chromosome 6A (6A: 545.832–574.480 Mb). Anchoring this minor QTL to the wheat physical map finds it to overlap with two previously identified QTL. The first is the Snn6 locus, which confers sensitivity to Tox6 (6A:574.222–606.980 Mb) (Gao et al. 2015), while the second is a seedling resistance QTL QSnl.ihar-6A (6A:579.126–583.269) (Arseniuk et al. 2004). Of note, a possible relationship between genetic loci controlling sensitivity to Tox3 and other effectors can be identified with the co-localisation of the Tox3 sensitivity locus Qsnb.fcu-4BL (4B: 608.252–657.496 Mb) (Phan et al. 2016) with the Tox5 sensitivity locus Snn5 (4B: 608.252–640.977 Mb) (Friesen et al. 2012). Direct links between the molecular pathways controlling different wheat sensitivity to P. nodorum effectors would intuitively make sense; however, whether this is true in practice remains to be further explored.
In summary, adult plant SNB resistance/susceptibility QTL on chromosomes 2A and 2D, and seedling infiltration resistance/susceptibility QTL on chromosome 7B previously identified in the UK ‘NIAB Elite MAGIC’ population were validated here in the German eight-parent BMWpop winter wheat MAGIC population. In the BMWpop, both haplotype effect at QSnb.nmbu-2A.1 and allele effect at QSnb.nmbu-5A.1 were significantly associated with field SNB susceptibility and significant across years, highlighting the robustness of these QTL. In addition, significant differences in SNB disease severity detected in 2016 between genotype groups showed evidence that the effect of these two field-relevant QTL was additive. As SNB resistance in the field is a complicated quantitative trait, validating field resistance QTL using an independent mapping panel provides robust evidence of the efficacy of target QTL in diverse genetic backgrounds. This knowledge should underpin efficient selection for beneficial SNB resistance alleles across multiple loci in wheat breeding programs, and will assist further research toward the identification of the functional allele(s) underlying these genetic loci.
References
Arseniuk E, Czembor PC, Czaplicki A, Song QJ, Cregan PB, Hoffman DL, Ueng PP (2004) QTL controlling partial resistance to Stagonospora nodorum leaf blotch in winter wheat cultivar Alba. Euphytica 137:225–231
Bhathal JS, Loughman R, Speijers J (2003) Yield reduction in wheat in relation to leaf disease from yellow (tan) spot and septoria nodorum blotch. Eur J Plant Pathol 109:435–443. https://doi.org/10.1023/A:1024277420773
Cockram J, Mackay I (2018) Genetic mapping populations for conducting high-resolution trait mapping in plants. Adv Biochem Eng Biotechnol 164:109–138. https://doi.org/10.1007/10_2017_48
Cockram J et al (2015) Fine-mapping the wheat Snn1 locus conferring sensitivity to the Parastagonospora nodorum necrotrophic effector SnTox1 using an eight founder multiparent advanced generation inter-cross population. G3 (Bethesda) 5:2257–2266
Corsi B et al (2020) Genetic analysis of wheat sensitivity to the ToxB fungal effector from Pyrenophora tritici-repentis, the causal agent of tan spot. Theor Appl Genet 133:935–950. https://doi.org/10.1007/s00122-019-03517-8
Czembor PC, Arseniuk E, Radecka-Janusik M, Piechota U, Słowacki P (2019) Quantitative trait loci analysis of adult plant resistance to Parastagonospora nodorum blotch in winter wheat cv. Liwilla (Triticum aestivum L.). Eur J Plant Pathol. https://doi.org/10.1007/s10658-019-01829-5
Downie RC et al (2018) Assessing European wheat sensitivities to Parastagonospora nodorum necrotrophic effectors and fine-mapping the Snn3-B1 locus conferring sensitivity to the effector SnTox3. Front Plant Sci 9:881. https://doi.org/10.3389/fpls.2018.00881
Duba A, Goriewa-Duba K, Wachowska U (2018) A review of the interactions between wheat and wheat pathogens: Zymoseptoria tritici, Fusarium spp. and Parastagonospora nodorum. Int J Mol Sci. https://doi.org/10.3390/ijms19041138
Faris JD et al (2010) A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc Natl Acad Sci U S A 107:13544–13549. https://doi.org/10.1073/pnas.1004090107
Ficke A, Cowger C, Bergstrom G, Brodal G (2018) Understanding yield loss and pathogen biology to improve disease management: Septoria Nodorum Blotch-a case study in wheat. Plant Dis 102:696–707. https://doi.org/10.1094/PDIS-09-17-1375-FE
Food and Agriculture Organization of the United Nations (2016). FAOSTAT [Online]. Available: https://www.fao.org/faostat/en/#home [Accessed 03/08 2019].
Fradgley N et al (2019) A large-scale pedigree resource of wheat reveals evidence for adaptation and selection by breeders. PLoS Biol 17:e3000071. https://doi.org/10.1371/journal.pbio.3000071
Francki MG (2013) Improving Stagonospora nodorum resistance in wheat: a review. Crop Sci 53:355–365. https://doi.org/10.2135/cropsci2012.06.0347
Francki MG, Walker E, McMullen CJ, Morris WG (2020) Multi-location evaluation of global wheat lines reveal multiple QTL for adult plant resistance to Septoria nodorum blotch (SNB) detected in specific environments and in response to different isolates. Front Plant Sci. https://doi.org/10.3389/fpls.2020.00771
Friesen TL, Faris JD (2012) Characterization of plant-fungal interactions involving necrotrophic effector-producing plant pathogens. In: Bolton MD, Thomma BPHJ (eds) Plant fungal pathogens: methods and protocols. Humana Press, Totowa, NJ, pp 191–207
Friesen TL, Meinhardt SW, Faris JD (2007) The Stagonospora nodorum-wheat pathosystem involves multiple proteinaceous host-selective toxins and corresponding host sensitivity genes that interact in an inverse gene-for-gene manner. Plant J 51:681–692. https://doi.org/10.1111/j.1365-313X.2007.03166.x
Friesen TL, Chu CG, Liu ZH, Xu SS, Halley S, Faris JD (2009) Host-selective toxins produced by Stagonospora nodorum confer disease susceptibility in adult wheat plants under field conditions. Theor Appl Genet 118:1489–1497. https://doi.org/10.1007/s00122-009-0997-2
Friesen TL, Chu C, Xu SS, Faris JD (2012) SnTox5-Snn5: a novel Stagonospora nodorum effector-wheat gene interaction and its relationship with the SnToxA-Tsn1 and SnTox3-Snn3-B1 interactions. Mol Plant Pathol 13:1101–1109
Gao Y, Faris JD, Liu Z, Kim YM, Syme RA, Oliver RP, Xu SS, Friesen TL (2015) Identification and characterization of the SnTox6-Snn6 interaction in the Parastagonospora nodorum-wheat pathosystem. Mol Plant Microbe Interact 28:615–625
Giraudoux P (2018). pgirmess: Spatial analysis and data mining for field ecologists. R package version 1.6.9. https://CRAN.R-project.org/package=pgirmess.
Hu W, He X, Dreisigacker S, Sansaloni CP, Juliana P, Singh PK (2019) A wheat chromosome 5AL region confers seedling resistance to both tan spot and Septoria nodorum blotch in two mapping populations. Crop J 7:809–818. https://doi.org/10.1016/j.cj.2019.05.004
Holloman DW (2015) Fungicide resistance: 40 years on and still a major problem. In: Ishii H, Holloman DW (eds) Fungicide resistance in plant pathogens. Springer, Japan
Huang BE, George AW (2011) R/mpMap: a computational platform for the genetic analysis of multiparent recombinant inbred lines. Bioinformatics 27:727–729. https://doi.org/10.1093/bioinformatics/btq719
International Wheat Genome Sequencing Consortium (IWGSC) et al. (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191 https://doi.org/10.1126/science.aar7191
Lin M, Corsi B, Ficke A, Tan KC, Cockram J, Lillemo M (2020a) Genetic mapping using a wheat multi-founder population reveals a locus on chromosome 2A controlling resistance to both leaf and glume blotch caused by the necrotrophic fungal pathogen Parastagonospora nodorum. Theor Appl Genet 133:785–808. https://doi.org/10.1007/s00122-019-03507-w
Lin M, Ficke A, Cockram J, Lillemo M (2020b) Genetic structure of the Norwegian Parastagonospora nodorum population. Front Microbiol 11:1280. https://doi.org/10.3389/fmicb.2020.01280
Liu Z, Friesen TL, Ling H, Meinhardt SW, Oliver RP, Rasmussen JB, Faris JD (2006) The Tsn1–ToxA interaction in the wheat–Stagonospora nodorum pathosystem parallels that of the wheat–tan spot system. Genome 49:1265–1273. https://doi.org/10.1139/g06-088
Liu Z et al (2009) SnTox3 acts in effector triggered susceptibility to induce disease on wheat carrying the Snn3 gene. PLoS Pathog 5:e1000581. https://doi.org/10.1371/journal.ppat.1000581
Liu Z et al (2012) The cysteine rich necrotrophic effector SnTox1 produced by Stagonospora nodorum triggers susceptibility of wheat lines harboring Snn1. PLoS Pathog 8:e1002467. https://doi.org/10.1371/journal.ppat.1002467
Liu Z et al (2015) Evaluation and association mapping of resistance to tan spot and Stagonospora nodorum blotch in adapted winter wheat germplasm. Plant Dis 99:1333–1341. https://doi.org/10.1094/PDIS-11-14-1131-RE
Mackay IJ et al (2014) An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: creation, properties, and validation. G3 (Bethesda) 4:1603–1610
Oliver RP, Friesen TL, Faris JD, Solomon PS (2012) Stagonospora nodorum: from pathology to genomics and host resistance. Annu Rev Phytopathol 50:23–43. https://doi.org/10.1146/annurev-phyto-081211-173019
Peters Haugrud AR, Zhang Z, Richards JK, Friesen TL, Faris JD (2019) Genetics of variable disease expression conferred by inverse gene-for-gene interactions in the wheat-Parastagonospora nodorum pathosystem. Plant Physiol 180:420–434. https://doi.org/10.1104/pp.19.00149
Phan HT, Rybak K, Furuki E, Breen S, Solomon PS, Oliver RP, Tan KC (2016) Differential effector gene expression underpins epistasis in a plant fungal disease. Plant J 87:343–354. https://doi.org/10.1111/tpj.13203
RStudio Team (2015). "RStudio: Integrated Development for R. RStudio, Inc., Boston, MA URL https://www.rstudio.com/.".).
Ruud AK, Lillemo M (2018) Diseases affecting wheat: Septoria nodorum blotch. In: Integrated disease management of wheat and barley. Burleigh Dodds Series in Agricultural Science. Burleigh Dodds Science Publishing Limited, Cambridge, UK, pp 109–144. https://doi.org/10.19103/as.2018.0039.06
Ruud AK, Windju S, Belova T, Friesen TL, Lillemo M (2017) Mapping of SnTox3-Snn3 as a major determinant of field susceptibility to Septoria nodorum leaf blotch in the SHA3/CBRD x Naxos population. Theor Appl Genet 130:1361–1374. https://doi.org/10.1007/s00122-017-2893-5
Ruud AK, Dieseth JA, Lillemo M (2018) Effects of three Parastagonospora nodorum necrotrophic effectors on spring wheat under Norwegian field conditions. Crop Sci 58:159–168. https://doi.org/10.2135/cropsci2017.05.0281
Ruud AK et al (2019) Genome-Wide Association Mapping of Resistance to Septoria Nodorum Leaf Blotch in a Nordic Spring Wheat Collection. The Plant Genome 12:1–15. https://doi.org/10.3835/plantgenome2018.12.0105
Scott MF, Ladejobi O, Amer S et al (2020) Multi-parent populations in crops: a toolbox integrating genomics and genetic mapping with breeding. Heredity. https://doi.org/10.1038/s41437-020-0336-6
Shi G et al (2016) The hijacking of a receptor kinase-driven pathway by a wheat fungal pathogen leads to disease. Sci Adv 2:e1600822. https://doi.org/10.1126/sciadv.1600822
Stadlmeier M, Hartl L, Mohler V (2018) Usefulness of a multiparent advanced generation intercross population with a greatly reduced mating design for genetic studies in winter wheat. Front Plant Sci. https://doi.org/10.3389/fpls.2018.01825
Stadlmeier M, Jorgensen LN, Corsi B, Cockram J, Hartl L, Mohler V (2019) Genetic dissection of resistance to the three fungal plant pathogens Blumeria graminis, Zymoseptoria tritici, and Pyrenophora tritici-repentis using a multiparental winter wheat population. G3 (Bethesda) 9:1745–1757
Tan KC, Waters ODC, Rybak K, Antoni E, Furuki E, Oliver RP (2014) Sensitivity to three Parastagonospora nodorum necrotrophic effectors in current Australian wheat cultivars and the presence of further fungal effectors. Crop Pasture Sci 65:150–158. https://doi.org/10.1071/Cp13443
Vleeshouwers VG, Oliver RP (2014) Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol Plant Microbe Interact 27:196–206. https://doi.org/10.1094/MPMI-10-13-0313-IA
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Wang S et al (2014) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796. https://doi.org/10.1111/pbi.12183
Wei J, Xu S (2016) A random-model approach to QTL mapping in multiparent advanced generation intercross (MAGIC) populations. Genetics 202:471–486. https://doi.org/10.1534/genetics.115.179945
Winfield MO et al (2016) High-density SNP genotyping array for hexaploid wheat and its secondary and tertiary gene pool. Plant Biotechnol J 14:1195–1206. https://doi.org/10.1111/pbi.12485
Zhang ZC, Friesen TL, Simons KJ, Xu SS, Faris JD (2009) Development, identification, and validation of markers for marker-assisted selection against the Stagonospora nodorum toxin sensitivity genes Tsn1 and Snn2 in wheat. Mol Breed 23:35–49. https://doi.org/10.1007/s11032-008-9211-5
Acknowledgements
We thank Cecilie Yri and Yalew Tarkegne (NMBU) for help with field phenotyping, and Prof. Richard Oliver (Curtin University) and Eiko Furuki (CCDM) for technical support and advice on effector usage.
Funding
Open Access funding provided by Norwegian University of Life Sciences. The research was undertaken within the ‘Efectawheat’ project supported within the framework of the 2nd call ERA-NET for Coordinating Plant Sciences, with funding to MIL from The Research Council of Norway grant NFR251894, to JC from Biotechnology and Biological Sciences Research Council (BBSRC) grant BB/N00518X/1, and to MS from the Deutsche Forschungsgemeinschaft (DFG, HA 5798/2–1), and to KT from Curtin University and the Grains Research and Development Corporation Grant CUR00023.
Author information
Authors and Affiliations
Contributions
MIL carried out glasshouse, field phenotyping and genetic analyses.VM provided the plant material. MS and VM developed the genetic analysis pipeline and helped with genetic analyses. JC and MOL gained project funding. KC supplied Tox3 effector. AF, JC and MOL supervised the research. MIL drafted the manuscript. All authors revised and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Communicated by Evans Lagudah.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lin, M., Stadlmeier, M., Mohler, V. et al. Identification and cross-validation of genetic loci conferring resistance to Septoria nodorum blotch using a German multi-founder winter wheat population. Theor Appl Genet 134, 125–142 (2021). https://doi.org/10.1007/s00122-020-03686-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-020-03686-x