Abstract
Key message
Using COS markers, the study reveals homeologous relationships between tetraploid Agropyron cristatum and bread wheat to support alien introgression breeding of wheat.
Abstract
Crested wheatgrass (Agropyron cristatum L. Gaertn.) is a wild relative of wheat that possesses many genes that are potentially useful in wheat improvement. The species comprises a complex of diploid, tetraploid and hexaploid forms. In this study, wheat–A. cristatum chromosome, telosome and translocation lines were used to characterize syntenic relationships between tetraploid A. cristatum and bread wheat. Prior to mapping COS markers, the cytogenetic stock lines were characterized for fertility and by FISH and GISH for karyotype stability. Out of 328 COS markers selected for the study, 279 consistently amplified products in tetraploid A. cristatum, and, out of these, 139 were polymorphic between tetraploid crested wheatgrass and wheat. Sixty-nine markers were found to be suitable for the detection of tetraploid A. cristatum chromosomes 1P–6P in wheat, ranging from 6 to 17 markers per chromosome. BLASTn of the source ESTs resulted in significant hits for 67 markers on the wheat pseudomolecules. Generally, COS markers of the same homeologous group were detected on similar arms in both Agropyron and wheat. However, some intragenomic duplications and chromosome rearrangements were detected in tetraploid A. cristatum. These results provide new insights into the structure and evolution of the tetraploid A. cristatum genome and will facilitate the exploitation of the wild species for introgression breeding of bread wheat.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The gene pool of bread wheat (Triticum aestivum L., 2n = 6x = 42, AABBDD) was narrowed down during thousands of years of domestication, cultivation and breeding. The dwindled genetic diversity hampers the development of cultivars with improved quality and tolerance to biotic and abiotic stresses. As wild crop relatives were not subjected to human selection, they exhibit large genetic variation and represent an attractive source of alleles and genes for crop improvement (Tanksley and McCouch 1997).
The genus Agropyron includes 10–15 species (Asay and Jensen 1996; Martín et al. 1999; Liu et al. 2010) and is a remarkable source of new gene variants for wheat improvement (Asay and Johnson 1990; Limin and Fowler 1990; Dong et al. 1992; Friebe et al. 1992). The most widespread species of the genus is A. cristatum (L.) Gaertn (Yang et al. 2014), also known as crested wheatgrass, which is a perennial, facultatively allogamic and autocompatible species. Agropyron cristatum contains the genome P in a series of diploid (2n = 2x = 14), tetraploid (2n = 4x = 28) and hexaploid (2n = 6x = 42) forms (Löve 1982, 1984; Dewey 1984; Li et al. 2007). The diploids are less common and distributed from Europe to Mongolia, whereas tetraploids are widespread, in central Europe, the Middle East and central Asia. Hexaploids are rare and are mainly found in Turkey, Iran and Georgia (Dewey and Asay 1982; Copete et al. 2018). The nature of the polyploid genome of A. cristatum and the origin of its P genome(s) have not yet been clarified. Schulz-Schaeffer et al. (1963) proposed the segmental allopolyploid nature of tetraploid and hexaploid A. cristatum, while other authors consider the tetraploid A. cristatum as autopolyploid, originating from the diploid A. cristatum (Taylor and McCoy 1973; Vogel et al. 1999; Zhao et al. 2017).
A number of agronomically important traits were identified in Agropyron spp., including resistance to pests and diseases such as barley yellow dwarf virus (Sharma et al. 1984; Shukle et al. 1987), wheat streak mosaic virus (Sharma et al. 1984; Brettell et al. 1988; Triebe et al. 1991), yellow rust, leaf rust and stem rust (Knott 1964, 1968; Cauderon and Rhind 1976; Whelan 1988; Friebe et al. 1992; Zhang et al. 2017), powdery mildew (Copete and Cabrera 2017) and tolerance to different abiotic stresses like cold (Limin and Fowler 1987), salinity (Dewey 1960, 1962; McGuire and Dvořák 1981; Forster et al. 1987; Littlejohn 1988) and drought (Dewey 1984) and genes affecting yield (Song et al. 2013). These traits would be desirable to transfer into wheat by interspecific hybridization. Interspecific hybridization is a promising tool to utilize the extant genetic diversity in wheat improvement through the chromosome-mediated transfer of useful agronomic traits (Feuillet et al. 2008). However, the crossability of diploid A. cristatum with wheat is low due to the disequilibrium in the endosperm balance number in the hybrid seeds (Chen et al. 1989), while hybridization between wheat and tetraploid A. cristatum can be easily done (Chen et al. 1989; Martín et al. 1999). As the crosses between wheat and hexaploid A. cristatum have not been reported yet, the tetraploid form is the most widely used crossing partner in the wheat-crested wheatgrass introgression breeding programs (Chen et al. 1989, 1994; Li et al. 1997, 1998, 2016; Soliman et al. 2007; Han et al. 2014; Ochoa et al. 2015).
In order to make A. cristatum genes accessible for wheat breeding programs, a set of wheat–A. cristatum (1P–6P) disomic addition lines has been developed by Chen et al. (1989, 1994) and Han et al. (2014) together with the production of ditelosomic additions 2PS, 2PL, 4PS, 5PL, 6PS or 6PL (Chen et al. 1989, 1994). Moreover, production of wheat–A. cristatum translocation lines was reported by Luan et al. (2010) and Song et al. (2013). In these studies, the chromosome 6P has been identified as the carrier of genes controlling yield components including the number of florets and kernels per spike (Luan et al. 2010) and of a locus conferring resistance to stripe rust (Zhang et al. 2017). Ochoa et al. (2015) developed another line, TH4, which carries a Robertsonian translocation involving the long arm of wheat chromosome 1B and the short arm of an unidentified tetraploid A. cristatum chromosome, with substantial resistance to leaf rust. The effectiveness of the gene transfer can be facilitated by high-throughput tools suitable for screening backcross populations for the presence of alien introgression lines with desired karyotype. On the other hand, the main selection tools are laborious cytogenetic methods, such as C-banding (Friebe et al. 1996), fluorescence in situ hybridization (FISH) (Rayburn and Gill 1985; Schwarzacher and Heslop-Harrison 2000; Schneider et al. 2005) and genomic in situ hybridization (GISH) (Schwarzacher et al. 1989; Le et al. 1989). The potential of FISH to identify particular chromosomes and their segments is also compromised by the lack of suitable probes, and the cytogenetic methods suffer from low sensitivity to detect small introgressed segments (Choi et al. 2009).
The efficiency of the gene transfer from wild relatives to wheat also depends on the homology between wheat and alien chromosomes. If collinearity between the donor and recipient genomes is broken down due to evolutionary chromosome rearrangements, homeologous recombination may result in progenies with nonbalanced genomes (Devos et al. 1993; Zhang et al. 1998). Altered structure of the donor chromosomes may interfere with meiotic recombination and hamper attempts to reduce the size of introgressed chromatin to eliminate undesirable traits (Nasuda et al. 1998). A further consequence of chromosome rearrangements, the genes on alien chromosome segments do not compensate for the loss of wheat genes and thus may negatively affect the agricultural performance of wheat–alien translocations. Polyploidization may induce genome rearrangements (Ma et al. 2004; Han et al. 2005, 2017; Zhang et al. 2013) and in case of A. cristatum Han et al. (2014) found that chromosome 6P of the tetraploid form differs from its wheat homeologous by large rearrangements. Later, Said et al. (2018) used single-gene FISH to investigate wheat–A. cristatum homeologous relationships and found several chromosomal rearrangements in diploid A. cristatum relative to wheat. However, the knowledge on the P genome structure of tetraploid A. cristatum remains poor and the mentioned observations underline the need for a structural comparison of P chromosomes in tetraploid A. cristatum with those of wheat.
Molecular markers have higher throughput than cytogenetic methods and capable of detecting small segments of A. cristatum chromatin; thus, they would facilitate the alien introgression breeding of wheat (Copete and Cabrera 2017). However, specific expressed sequence tag (EST) markers have been developed only for chromosomes 6P (Cheng et al. 2012) and sequence-tagged site (STS) markers for 7P (Lu et al. 2016) of tetraploid A. cristatum. Thus, the low coverage of P genome with small number of molecular markers limits the efficiency of introgression breeding and the investigation of wheat–tetraploid A. cristatum homeologous relationships. The chromosome-based genome assembly is still not available in species with large and complex genomes such as A. cristatum with a 1C genome size of 6352 Mbp (Said et al. 2018), a fact that strongly limits the development of DNA markers for the P genome. An alternative strategy is exploiting molecular markers from genetically related species and their assignment to individual chromosomes of the donor genome using alien introgression lines (Said and Cabrera 2009; Cherif-Mouaki et al. 2011; Said et al. 2012; Copete and Cabrera 2017).
Conserved orthologous set (COS) genes (> 1000) are well preserved throughout evolution in both sequence and copy number between wheat and the model species rice and Brachypodium (Quraishi et al. 2009). COS markers were designed over the exon–intron boundaries of orthologous genes. As introns sequences are less conserved than exons (Yu et al. 2005), COS markers are potentially polymorphic. The polymorphism can be detected as a difference in the size of PCR products amplified from wheat and related species. In agreement with this, COS markers are highly transferable between species such as rice, wheat, maize, sorghum, barley and several species of Aegilops (Parida et al. 2006; Burt and Nicholson 2011; Howard et al. 2011; Molnár et al. 2013, 2016). Wheat-specific COS markers are also transferable to diploid wheatgrass species, such as Thinopyrum bessarabicum (Savul.&Rayss) A. Löve, Pseudoroegneria spicata (Pursh) A. Löve and A. cristatum (Linc et al. 2017). Because this marker system is based on orthologous genes, COS markers enable the comparison of orthologous regions on the chromosomes of related species. Using COS markers on wheat–Aegilops disomic addition lines, Molnár et al. (2013) assigned a total of 132 and 156 loci to the U and M genome chromosomes of Aegilops and investigated the homeologous relationships between the U and M genomes of Aegilops and wheat. Later, Gaál et al. (2018) assigned sixty COS markers to the E-genome chromosomes of Thinopyrum elongatum revealing several chromosomal rearrangements relative to wheat.
Motivated by the need to improve the efficiency of the chromosome-mediated gene transfer from tetraploid A. cristatum into wheat, the aim of the present study was to increase the number of markers suitable to detect P chromosomes in the wheat genetic background and to study orthologous relationships between the chromosomes of A. cristratum and bread wheat. Using PCR with DNA from a wheat–A. cristatum disomic addition lines 1P, 2P, 3P, 4P, 5P and 6P, and wheat–A. cristatum telosome addition lines 2PS, 2PL, 4PS, 5PL, 6PS and 6PL (Chen et al. 1989, 1994), we assigned wheat-specific COS markers to the chromosomes and chromosome arms of Agropyron. Sequence similarity search of the source ESTs of the Agropyron specific COS markers on the wheat pseudomolecules has also been done to investigate wheat–Agropyron genome relationships.
Materials and methods
Plant material
The seeds of diploid A. cristatum cv. Parkway (2n = 2x = 14, PP), accession number PI 415799 were provided by Dr Joseph Robins (ARS Forage and Range Research Laboratory, USDA, Logan, USA). The seeds of tetraploid A. cristatum (2n = 4x = 28, PPPP) accession PI 222957 were obtained from the USDA genebank (https://npgsweb.ars-grin.gov/gringlobal/search.aspx). The seeds of parental wheat cv. Chinese Spring (CS) together with the seeds of CS-A. cristatum chromosome addition lines (CS-P) carrying chromosomes 1P, 2P, 3P, 4P, 5P and 6P (CS-1P–6P), telosome addition lines (CS-PS, PL) for chromosome arms 2PS, 2PL, 4PS, 5PL, 6PS and 6PL were produced by Chen et al. (1989, 1994) were also used in the present study as well as the CS-A. cristatum Robertsonian translocation line (CST-P) involving the long arm of wheat chromosome 1B and the short arm of an unidentified chromosome from tetraploid A. cristatum (Ochoa et al. 2015). The new CS-A. cristatum telosome addition line 3PS (CS-3PS) was developed in the present study at the Institute of Experimental Botany (Olomouc, Czech Republic) by successive selfing the line CS-3P possessing one 3P chromosome and one P telosome and selecting for the genotypes carrying mono- or ditelosomic 3PS (Fig. 1).
Karyotype stability, fertility and morphological characteristics
Before the COS marker analysis, all wheat–A. cristatum chromosome and telosome addition lines and the translocation line were characterized for fertility and karyotype stability during three successive generations. Fertility and spike morphological characteristics were estimated from five spikes for the following traits: spikelets per spike, seeds per spike and spike length. An analysis of variance was carried out, and a mean values comparison was accomplished using the least significant difference method (P ≤ 0.05). Statistical analysis was performed with Minitab 18 (www.minitab.com). Karyotype stability of the wheat–A. cristatum lines was evaluated by chromosome counting and genomic in situ hybridization (GISH) as detailed in Szakács and Molnár-Láng (2010). Chromosome compositions of the wheat–A. cristatum lines over the generations were expressed in percent (%): the number of plants with a specific chromosome composition divided by the total number of plants analyzed from a specific line and multiplied by 100.
Preparation of probes for FISH
A probe for A. cristatum tandem repeat ACRI_CL78 (Said et al. 2018) was labeled by PCR with digoxigenin-dUTP (Roche, Mannheim, Germany) using diploid A. cristatum cv. Parkway DNA as a template. Biotin-dUTP (Roche) or digoxigenin-dUTP labeled probe for 5S rDNA was prepared according to Fukui et al. (1994) using rice DNA as a template for PCR. The plasmid pTa71 (45S rDNA) containing a 9-kb fragment from bread wheat with 18S-5.8S-26S rDNA and intergenic spacers (Gerlach and Bedbrook 1979) and genomic DNA of tetraploid A. cristatum PI 222957 were labeled with either biotin or digoxigenin by nick translation using standard kits (Nick Translation Mix, Roche) following the manufacturer’s instructions.
Mitotic chromosome preparation
Seeds were germinated on moistened filter paper in a glass Petri dish in the dark at 25 °C for 3–4 days. Root tips were transferred to distilled water and incubated overnight at 1 °C in a box filled with ice water. Subsequently, the root tips were fixed in ice-cold 90% acetic acid for 10 min followed by three washes in 70% ethanol and stored in 70% ethanol at − 20 °C. Chromosome preparations were prepared using the drop technique according to Kato et al. (2004, 2006), with minor modifications as described in Danilova et al. (2012).
Fluorescence in situ hybridization
Labeled probes for FISH and GISH were localized following the protocols of Cabrera et al. (2002) and Said et al. (2018) with modifications. Briefly, digoxigenin-labeled probes were detected using anti-digoxigenin fluorescein isothiocyanate (Roche). Biotin-labeled probes were detected with Cy3-conjugated streptavidin (Invitrogen, Life Technologies, Carlsbad, USA). The hybridization mixture (total volume = 10 μl/slide) contained 50 ng labeled probe DNA, 50% v/v formamide, 2 × SSC (0.15 mol/l NaCl plus 0.015 mol/l sodium citrate), 10% w/v dextran sulfate, 0.4 μg salmon sperm DNA and 0.1% w/v sodium dodecyl sulfate. In the case of GISH, 5 μg wheat genomic DNA was included in the hybridization mix as blocking DNA. The chromosomes and probes were denatured together at 80 °C for 3 min under high moisture conditions. The hybridization was carried out overnight at 37 °C. The slides were washed, the hybridization sites were detected, and chromosomes were mounted and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) in Vectashield media (Vector Laboratories, Burlingame, USA).
Microscopy, software, signal capture and image analysis
Chromosome preparations were examined using an Axio Imager Z.2 Zeiss microscope (Zeiss, Oberkochen, Germany) equipped with a Cool Cube 1 (MetaSystems, Altlussheim, Germany) camera and appropriate optical filter sets. The signal capture and image processing were performed using ISIS software (MetaSystems). The final image adjustment was done in Adobe Photoshop CS5 (Adobe Systems Incorporated, San Jose, USA).
COS marker analysis
Genomic DNA was extracted from young leaves of wheat–A. cristatum chromosome and chromosome arm addition and translocation lines, from the diploid and tetraploid A. cristatum accessions and from bread wheat cv. Chinese Spring, using a QuickGene Mini 80 (FujiFilm, Tokyo, Japan) with a QuickGene DNA tissue kit (FujiFilm) according to the manufacturer’s instructions and was used as a template for PCR. Primers for 328 COS markers covering wheat homeologous groups 1–7 (group-1: 76, group-2: 16, group-3: 23, group-4: 120, group-5: 65, group-6: 15 and group-7: 13) were chosen from publicly available marker collections (Quraishi et al. 2009; Howard et al. 2011). Primer sequences for these markers and annealing temperature (Ta) are summarized in Supplementary Data S1. PCR was performed in 12 µl reaction volumes as described by Molnár et al. (2014, 2016) using a touchdown reaction profile: 94 °C 2 min, 10 cycles of 94 °C 0.5 min, Ta + 5 °C 0.5 min and decreased by 0.5 °C increments for every subsequent set of cycles, 72 °C 1 min, 30 cycles of 94 °C 0.5 min, Ta °C 0.5 min, 72 °C 1 min, hold at 72 °C 2 min in an Eppendorf Mastercycler (Eppendorf, Hamburg, Germany). PCR products of the 112 markers, which were analyzed in the Agricultural Institute, Hungarian Academy of Sciences (Martonvásár, Hungary), were separated by a Fragment Analyzer Automated CE System equipped with a 96-Capillary Array Cartridge with an effective length of 33 cm (Advanced Analytical Technologies, Ames, USA), and the results were analyzed and visualized by PROsize v2.0 (Advanced Analytical Technologies). The products of the remaining 216 markers, which were analyzed at the University of Córdoba, were separated on 2.5% agarose gels along with the O’RangeRuler™ 50 bp DNA size marker (Fermentas, Vilnius, Lithuania) as described by Nagy et al. (2006). The patterns were documented and analyzed using a GeneGenius gel documentation system (Syngene, Cambridge, UK). The PCR products were considered as polymorphic if difference in the product size between wheat and Agropyron was ≥ 3 bp.
Sequence analysis
The orthologous relationship between the chromosomes of wheat and A. cristatum has been investigated from the genomic perspective of wheat. As a first step for the construction of the physical maps, the EST source sequences (Supplementary Data S2) were used as queries in BLASTn searches against the wheat reference pseudomolecules (Consortium (IWGSC) et al. 2018) to identify the start positions (bp) of the ESTs. Throughout the study, BLAST hits with E values smaller than 2.8e−08, identity % > 58.44 and alignment length > 100 bp were considered significant. The genomic start positions in bp of the best hits in wheat pseudomolecules (Supplementary Data S3) were used to construct physical maps of the polymorphic COS markers. The centromere positions for each wheat chromosome were obtained from the wheat reference genome sequence (Consortium (IWGSC) et al. 2018). The length in bp of wheat pseudomolecules, as well as the start genomic positions of the ESTs, was converted to pixels. Then, the BLASTn searches data were used to construct a physical map for each of the wheat chromosomes showing the position of the source EST of the COS markers assigned to Agropyron chromosomes. The wheat chromosomes were colored with six different colors representing the homeologous groups (1–6) of both species: wheat and A. cristatum. However, the marker name background was colored with colors correspond to their arm location on A. cristatum chromosomes as described by Molnár et al. (2016). Dark and light colors of the chromosomes and the background color of the marker name represent the short and long arms of wheat chromosomes and the arm location on A. cristatum chromosomes, respectively. The physical maps of the COS markers were designed using custom-made software.
Accession numbers
Plant material; diploid A. cristatum cv. Parkway (2n = 2x = 14, PP), accession number PI 415799 and tetraploid A. cristatum (2n = 4x = 28, PPPP) accession PI 222957 are available at the USDA genebank (https://npgsweb.ars-grin.gov/gringlobal/search.aspx). Wheat–A. cristatum chromosome additions (1P, 2P, 3P, 4P, 5P and 6P) and telosome additions (2PS, 2PL, 3PS, 4PS, 5PL, 6PS and 6PL) as well as a wheat–A. cristatum 1PS·1BL translocation line are available upon request: Mahmoud Said (said@ueb.cas.cz), Institute of Experimental Botany, Šlechtitelů 31, CZ-78371 Olomouc, Czech Republic.
Results
Karyotype stability of the wheat–A. cristatum chromosome addition and translocation lines
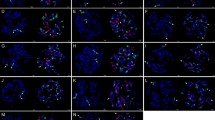
Karyotype stability was observed in a majority of the addition and translocation lines (Supplementary Table S1). Complete stability was observed in CS-6PL addition lines and CST-P translocation lines with 100% maintenance of the disomic state of the alien chromatin. The second greatest stability was observed for addition lines CS-1P, CS-4P, CS-5P, CS-6P, CS-5PL and CS-6PS, where more than 90% of the progeny retained the disomic state. The CS-2P, CS-2PL, CS-3PS and CS-4PS addition lines were relatively stable, as more than 77% disomic progeny plants were identified. Furthermore, the CS-2PS addition line was unstable, with 47% ditelosomic and 10% monotelosomic progeny, while the remaining 43% plants did not retain any alien chromatin. Line CS-3P was the most unstable, as a high proportion of plants (74%) did not retain any alien chromatin, 17% of the progenies were monotelosomic, and 9% of the plants retained one chromosome 3P in addition to a chromosome arm later identified by FISH as 3PS. The 3PS chromosome arm was also detected in a new ditelosomic addition line CS-3PS, which was generated during the course of the study. This line showed high stability, where 77% of progenies maintained the ditelosomic state. Although the CS-6PS addition line was considered highly stable with 90% ditelosomic plants in the progeny, 5% of plants carried isochromosome 6PS and the remaining 5% were monotelosomic as revealed by GISH. The wheat–A. cristatum lines with disomic and ditelosomic additions confirmed by GISH (Fig. 2, Supplementary Fig S1) were selected for COS marker analyses.
GISH on mitotic metaphase plates of wheat–A. cristatum lines using genomic DNA from tetraploid A. cristatum (green). a Chromosome 4P disomic addition b chromosome arm 4PS ditelosomic addition and c homozygous translocation 1PS·1BL lines. Chromosomes were counterstained by DAPI (blue). Please refer to the online version for the high-resolution color figure
We observed two karyotypes in the progenies of line CS-3P, one retaining one chromosome 3P and one arm 3P, and the other karyotype of a new line possessing two telosomes (Fig. 1). Based on the FISH pattern of the tandem repeat ACRI_CL78 (Said et al. 2018), the telosomes were identified as a homologous pair 3PS of a new ditelosomic addition line CS-3PS (Supplementary Fig S2). Furthermore, the telosomes of the new line CS-3PS had similar FISH pattern to the short arm of chromosome 3P of line CS-3P (Supplementary Fig S2). The results were also confirmed by the comparison of the FISH pattern of the tandem repeat on the short arm of chromosome 3P and the telosome in line CS-3P possessing one chromosome 3P and one arm (Supplementary Fig S2). The unidentified tetraploid A. cristatum chromosome short arm translocated to wheat chromosome arm 1BL (Ochoa et al. 2015) in the translocation line CST-P was identified in the present work by FISH as 1PS based on the molecular karyotype of A. cristatum (Said et al. 2018), as it is possible to distinguish between chromosomes 1P and 5P. Although both are characterized by 45S rDNA signals at the terminal position of the short arms, chromosome 5P has a 5S rDNA locus at the subterminal position of the short arm. This was also confirmed by comparing the distribution patterns of 45S rDNA on this chromosome arm in the translocation line with the patterns of 5S and 45S rDNA on tetraploid A. cristatum PI 222957 (Supplementary Fig S3), which was used by Ochoa et al. (2015) to develop the translocation. Consequently, based on these observations, the translocation line (CST-P) was renamed to CST-1PS·1BL.
The new wheat–A. cristatum CS-3PS ditelosomic line and the translocation line CST-1PS·1BL were also involved in the COS marker study (Fig. 2, Supplementary Fig S1). Because the whole 3P chromosome was not represented in the set of wheat–A. cristatum disomic addition lines, we used line CS-3P which possesses one chromosome 3P and one telosome 3PS (Supplementary Fig S1).
Fertility and morphological traits
The observations on fertility and spike morphology are summarized in Supplementary Table S2 and Fig. 3. All lines were fertile and vigorous over the generations, both in a greenhouse and under field conditions. The lines differed in spike morphology in terms of color, size and shape, and statistically significant differences were found between the lines for the evaluated characters. In particular, translocation line CST-1PS·1BL had a significantly longer spikes compared to CS, while the CS-1P addition line had the shortest spike with approximately half the spike length of CS. Line CS-1P had significantly fewer spikelets per spike compared to the remaining lines, while CS-2PL showed the significantly highest value for this trait. With respect to seed number per spike, CS-6P had a greater mean value, but the difference was not significant when compared to CS, CS-4PS and CS-6PL. Line CS-2P had the lowest number of seeds per spike, but the difference was not significant compared to CS-1P and CS-4P. In this study, all wheat–A. cristatum lines yielded awnless spikes, except for line CS-2P which had awned spikes, and fewer and shorter awns were also observed on the upper spikelets of CS-2PS and CS-2PL (Fig. 3).
Assignment of COS markers to P chromosomes
The confirmation of the presence of chromatin originating from tetraploid A. cristatum in wheat–A. cristatum addition and translocation lines made them suitable for the subsequent COS markers analysis. Out of the 328 markers tested for transferability to tetraploid A. cristatum (Table 1), 279 (85.1%) consistently amplified products in tetraploid A. cristatum PI 222957, and, out of these, 139 (49.8%) were polymorphic between tetraploid A. cristatum and wheat (CS) (Fig. 4 and Table 1). The highest level of polymorphism (90.0–90.9%) was observed for the markers of wheat chromosome groups 3 and 6, while the markers of wheat chromosome groups 1 and 4 showed a relatively low level of polymorphism (32.0–35.0%). Using wheat–A. cristatum disomic- and ditelosomic addition lines, sixty-nine out of the 139 polymorphic markers were assigned to the P genome chromosomes. Because some markers were assigned to more than one P chromosome (Table 2), the total number of markers (69) assigned to tetraploid A. cristatum chromosomes was different from the sum (78) of the specific markers per P chromosome (no. of markers/no. of PCR amplicons per chromosome: 1P: 11/15; 2P: 6/7; 3P: 10/14; 4P: 19/24; 5P: 21/24; 6P: 11/13).
PCR amplification profiles used for the location of COS molecular markers on chromosomes 1P, 4P and 5P. a, b c737237 and c743346 mapped on the short and long arms of chromosome 1P, respectively; c, d c763059 and c739175 mapped on the short and long arms of chromosome 4P, respectively; e, f c728036 and c762245 mapped on the short and long arms of chromosome 5P, respectively
The availability of CS-A. cristatum ditelosomic lines provided the opportunity to locate COS markers on chromosome arms. Because ditelosomic lines for the short and long arms were available only for chromosomes 2P and 6P, only one of the two arms could be checked for chromosomes 3P, 4P and 5P and for 1P using the CST-1PS·1BL translocation. Therefore, if the PCR results were negative in the available telosomic line and positive in the whole chromosome addition line, we concluded that the COS marker was located on the opposite arm. In this manner, sixty-seven out of the sixty-nine markers were assigned to chromosome arms of the tetraploid A. cristatum (Table 2). We failed to map the remaining two markers to particular chromosome arms. The tetraploid Agropyron chromosome-specific markers showed a significant level of length polymorphism (3–558 bp, mean 54.59 bp) between wheat and the parental tetraploid A. cristatum genotype represented by the wheat–A. cristatum addition and ditelosomic lines. Therefore, they were considered suitable for marker-assisted selection of new wheat–Agropyron introgression lines in prebreeding programs. In this study, 90 polymorphic loci of 69 markers (1–3 loci, 1.30 loci per marker) covering from one to six homeologous groups of the P genome were found to be suitable for high-throughput detection of tetraploid A. cristatum chromosomes.
Wheat–A. cristatum homeology at the chromosome level
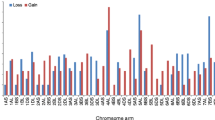
To investigate wheat–A. cristatum homeologous relationships at the chromosome level, the source ESTs of the 69 polymorphic COS markers were BLASTed to the sequences of the wheat chromosomes (Consortium (IWGSC) et al. 2018). Sixty-seven markers showed hits on wheat pseudomolecules, and two markers (TR451, TR430) gave no hits. Seven markers (TR73, TR85, c750766, c756425, c746156, c759134 and TR764) were excluded from the subsequent analysis because the alignment length was below the threshold (100 bp). For the remaining markers, the start positions of the alignments of the best hits on the A, B and D genomes were extracted to produce a physical map from the perspective of the wheat genome (Supplementary data S3). In the map, the chromosomal locations of the markers in tetraploid A. cristatum were visualized by different colors of the marker name background, which provides an overview of the wheat–A. cristatum genome relationships from the perspective of the wheat genome (Fig. 5). In the physical map, the coverage of wheat chromosome groups 2, 3 and 6 with COS markers was smaller (6–7 markers per chromosome group) than wheat chromosome group-1 (10 markers) and wheat chromosome groups 4 and 5 (15–16 markers per chromosome).
Genome relationships defined by polymorphic COS markers in wheat and A. cristatum from the genomic perspective of wheat homeologous groups 1–6 (Group-7 was omitted from this study due to the unavailability of a wheat–A. cristatum 7P addition line). The name of the markers is on the right, while their physical positions (kb) on the wheat pseudomolecules are on the left. The wheat chromosomes were colored with six different colors representing homeologous groups (1–6). The background color of the marker name indicates the chromosomal location of the marker in A. cristatum. If the chromosome and the background of the marker name have the same color, the marker assigned to the same homeologous group chromosome in wheat and A. cristatum. For example, the group 1 chromosomes of wheat are red and the background of the nine marker names is red, indicating that these nine markers are assigned to the group 1 chromosomes in wheat and Agropyron (1ABD and 1P). However, the turquoise color indicates that the marker c757404, which is located on the 1A, 1B and 1D at 127075.2, 177835.9 and 116003 kb, respectively, in wheat, was assigned to 4P chromosome of A. cristatum. Dark and light colors indicate short (S) and long (L) arms, respectively. The centromere positions (arrows) for each wheat chromosome were determined from the wheat reference genome sequence (Consortium (IWGSC) et al. 2018). Please refer to the online version for higher resolution color figure
Most of the markers (48) located on the same homeologous (H) chromosome group in tetraploid Agropyron as in wheat, while seven markers were assigned to nonhomeologous (N) P genome chromosomes. The remaining five markers have duplicated loci as they were assigned to both homeologous and nonhomeologous (H/N) chromosomes of A. cristatum.
The marker c757404, which is located on the interstitial part of the short arm of wheat chromosome group-1, was assigned to the short arm of 4P. The marker TR72, specific to the subtelomeric region of wheat chromosome group-2 short arms, was located at 3P. The marker TR4, specific to the subtelomeric region of short arms of wheat chromosome group-3, was assigned to 5PL, while TR390, specific to the long arms of the interstitial part of wheat chromosome group-3, was located at 1P. On wheat chromosome group-4, two markers (TR118 and TR133) were mapped to the 5P chromosome and one marker c797119 was assigned to 6P. The wheat chromosome group-5 markers TR390, TR759 and TR471 were detected on chromosomes 1P, 6P and 3P, respectively, while the duplicated locus TR636, which is located on wheat chromosome groups 2 and 6, was identified on 2P.
Inter- and intrachromosomal rearrangements in the P genome
As shown in Table 2 and in the physical map (Fig. 5), 53 (48 H and 5 H/N) out of 60 (88.3%) COS markers showed synteny between the bread wheat and tetraploid A. cristatum genomes as they were detected on the same homeologous chromosome groups, with 46 (76.7%) of them at the same short or long arm in tetraploid Agropyron and wheat. However, the remaining seven (11.7%) COS markers mapped to nonhomeologous chromosomes and thus revealed structural differences between the chromosomes. Ten markers (16.7%) indicated intrachromosomal rearrangements as these markers were specific for the same homeologous chromosomes in tetraploid Agropyron as in wheat, but they located at the opposite arms. These kinds of intrachromosomal rearrangements were found on A. cristatum chromosomes 1P, 3P, 4P, 5P and 6P (Supplementary Table S3 and Fig. 5). Furthermore, the chromosomal location of other COS markers revealed some intragenomic duplications in tetraploid A. cristatum relative to wheat (Table 2 and Fig. 5). Three duplications were detected by markers specific to wheat chromosome group-2. Loci for marker 2R on the short arms of wheat chromosome group-2 detected a duplication of 2PL/5PL, while markers from long arms detected two duplications 2PL/6PS and 2PL/5PL/6PS were found by the markers c744070 and 2N, respectively. One duplication, 1PL/3PL/4PL/6PL, was detected by the marker c803223, which is specific for the telomeric region of wheat chromosome group-3, while a 4PL/6PS duplication was found by the marker BE426214, which is specific for the long arms of wheat chromosome group-6.
Discussion
The present study significantly increased the number of chromosome-specific molecular markers suitable for improving the selection throughput of wheat–A. cristatum introgression lines with requested alien chromatin.
Before using the wheat–Agropyron lines for structural genomic studies, cytogenetic control is needed because the elimination of alien chromosomes from wheat–alien genetic stocks is a well-known feature (Taketa et al. 1995; Molnár-Láng et al. 2005; Szakács and Molnár-Láng 2010). In this work, different levels of instability were observed in wheat–A. cristatum lines, particularly in CS-3P addition line, where the Agropyron chromosome was not observed in disomic form. These results are in line with previous studies demonstrating that wheat cytogenetic stocks carrying chromosome 3 from wild relatives of wheat are difficult to maintain in disomic form (Miller et al. 1982; Said et al. 2012). A recent study using 3D-FISH on wheat–rye introgression lines showed that incorrect arrangement in the nuclei at the beginning of meiosis may prevent the migration of alien introgressed chromosomes into the telomere bouquet, thereby preventing synapsis and chiasma formation, and leading to their gradual elimination (Perničková et al. 2019). A similar study in the future may help to understand the cytogenetic background of the high eliminating rate of 3P chromosomes from the wheat–A. cristatum addition line.
The cytogenetic analysis allowed to identify a new 3PS ditelosomic addition line using the tandem repeat probe ACRI_CL78 which has a subterminal signal diagnostic for 3PS (Said et al. 2018). By the use of 5S rDNA and 45S rDNA probes, we also identified the P genome chromosome arm involved in the wheat–A. cristatum centric fusion CST-P·1BL developed by Ochoa et al. (2015) as 1PS according to the A. cristatum karyotype developed by Said et al. (2018). The identification of A. cristatum chromatin in these two lines enabled the assignment of markers to 1PS and 3PS.
Spike architecture and fertility are important components contributing to the yield of bread wheat. Our results indicating that chromosomes 2P, 4P and 6P carrying genes contributing positively to the number of spikelets per spike which is accompanied by a reasonable fertility (2.76–2.81) in CS-6P, CS-6PL and CS-4PS resulting in the same level of seeds per spike as the parental wheat genotype CS. These results support the previous observations on the positive effect of chromosome 6P on grain yield of bread wheat reported by Wu et al. (2006), Ye et al. (2015) and Zhang et al. (2015). The effect of Agropyron genes on the spike architecture of wheat might be more pronounced after shortening the size of introgressed 4P or 6P chromatin by induced homeologous recombination or by random chromosome breakage (Shi et al. 2005; Endo 2015; Martín et al. 2017). A marker-based system would also make more effective the selection of genotypes with shorter chromatin.
Easy to use, chromosome-specific molecular markers are a prerequisite for increasing the selection throughput of wheat–alien introgression lines with desirable karyotypes. The co-dominant COS markers meet this criteria, as they amplify specific PCR products from wheat and the introgressed chromatin is suitable for discrimination between heterozygotes and homozygotes in F2 and BC populations. One of the two primers is specific for the conserved exon sequences; thus, the markers have theoretically high transferability between the species, while the other primer is positioned, so that the product covers the intron. As intron sequences are less conserved than exons, the size-polymorphic product obtained in wheat–alien introgression lines reflects the species specific genetic variations relative to wheat.
We observed 85.1% transferability of COS markers between wheat and tetraploid A. cristatum, which is less than those reported by Linc et al. (2017), who found 92.1% transferability of COS markers between wheat and diploid A. cristatum. However, it is higher than those found by Copete and Cabrera (2017), who obtained 68.2% transferability of wheat chromosome groups 2 and 6 specific COS markers between wheat and tetraploid A. cristatum. In comparison with other marker systems, the transferability of wheat-specific COS markers can be higher than EST-STS markers, it can be comparable with those of EST-SSRs and can be lower than those of PLUG markers as it was reported for Aegilops caudata (Gong et al. 2017) and Aegilops searsii (Gong et al. 2016).
In this work, we significantly increased the number of PCR-based markers available for detection of chromosomes 1P–6P of tetraploid A. cristatum and their arms in the wheat background. We found that out of the 279 COS markers producing amplicons in A. cristatum, 139 (49.8%) showed size-polymorphic product between wheat and tetraploid A. cristatum. A similar range of size polymorphism (54.27%) was reported by Han et al. (2014) for EST-SSR markers between the wheat cultivar ‘Fukuhokomugi’ and tetraploid A. cristatum genotype Z559. Interestingly, the same work showed a much smaller percentage of size polymorphism (36.95%) for genomic SSR markers. Our work significantly expanded the number of P chromosome-specific markers by identifying the chromosomal locations of 69 COS markers covering the 1P–6P chromosomes of tetraploid A. cristatum. These polymorphic markers are considered potentially useful to follow tetraploid A. cristatum chromosomes in bread wheat backgrounds during prebreeding programs.
Tetraploid A. cristatum is considered autopolyploid, and the markers are specific for its both subgenomes. However, intraspecific diversity may cause presence/absence or size variation in the amplified loci. In a previous work, Linc et al. (2017) investigated genetic diversity using COS markers in perennial grass species, including two accessions of diploid A. cristatum. Among the 96 markers gave amplicon on A. cristatum, 46 (76 loci) were nonpolymorphic, while 50 markers (106 loci) were polymorphic between the two accessions of A. cristatum.
Homeologous relationships between wheat and related species provide important information for the targeted development of markers specific to alien chromosome regions potentially responsible for important agronomic traits as demonstrated for Aegilops ventricosa (Burt and Nicholson 2011). The present study revealed close homeologous relationships between the chromosome arms of bread wheat and tetraploid A. cristatum as 48 markers were located on the same homeologous chromosomes of wheat as those of Agropyron. However, this arm-level homeology was perturbed in some loci as one and one markers indicated 1ABDS-4P, 2ABDS-3P, 3ABDS-5P, 3ABDL-1P, 4ABD-5P, 4ABD-6P, 5ABD-1P, 5ABD-3P, 5ABD-6P and 6ABDS-2P relationships, while others indicated 2P/5P or 2P/5P/6P duplications. These results agree well with those of Zhou et al. (2018) who used wheat 660 k SNP array to genotype a diploid A. cristatum × Allium mongolicum segregating population and also found the P genome of Agropyron is generally collinear, but some rearrangements exist relative to wheat genomes. Some markers are also indicated intrachromosomal rearrangements, such as peri- and paracentric inversions, on the P genome chromosomes, which is in line with the cytogenetic observations of Said et al. (2018), who used single-gene FISH and detected rearrangements on chromosomes 2P–7P, including peri- and paracentric inversions on 4P and 6PL, respectively, of diploid A. cristatum relative to wheat. Our results based on the chromosomal location of COS markers may indicate that chromosomal inversions are more abundant in the P genome of tetraploid A. cristatum than those of the diploid form, which could be the consequence of the polyploidization-induced genome reorganization (Ma et al. 2004; Han et al. 2005, 2017; Zhang et al. 2013). In line with these studies, Said et al. (2019) observed differences in the FISH signal pattern between chromosomes flow sorted from the diploid and tetraploid accessions, and the differences were more pronounced on the chromosomes flow sorted from tetraploid A. cristatum. A recent development in the flow-cytometric sorting of P genome chromosomes from the wheat–A. cristatum addition and ditelosomic lines opens the way for shot gun sequencing and high-resolution structure analysis of A. cristatum chromosomes (Said et al. 2019).
Conclusions
In the present study, a set of COS markers was successfully assigned to the chromosomes and chromosome arms of the P genome of tetraploid A. cristatum, which is the only form of this wild species suitable for chromosome-mediated gene transfer to bread wheat. Our results revealed the genome structure and the homeologous relationships of this species relative to wheat, which could help us to understand the evolution of species from the Triticeae tribe, open the door for genome analysis and support the use of this important wild gene source in wheat breeding.
Author contribution statement
AC, JD, JV, IM and MS conceived the project. ACP, EG, JV, IM and MS performed the experiments and drafted the manuscript; all authors contributed to the manuscript writing and approved the final version.
References
Asay KH, Jensen KB (1996) Wheatgrasses 1. American Society of Agronomy, Crop Science Society of America, Soil Science Society of America Agronomy Monograph no 34, pp 691–724. https://doi.org/10.2134/agronmonogr34.c22
Asay KH, Johnson DA (1990) Genetic variances for forage yield in crested wheatgrass at six levels of irrigation. Crop Sci 30:79–82. https://doi.org/10.2135/cropsci1990.0011183X003000010018x
Brettell RIS, Banks PM, Cauderon Y et al (1988) A single wheatgrass chromosome reduces the concentration of barley yellow dwarf virus in wheat. Ann Appl Biol 113:599–603. https://doi.org/10.1111/j.1744-7348.1988.tb03337.x
Burt C, Nicholson P (2011) Exploiting co-linearity among grass species to map the Aegilops ventricosa-derived Pch1 eyespot resistance in wheat and establish its relationship to Pch2. Theor Appl Genet 123:1387–1400. https://doi.org/10.1007/s00122-011-1674-9
Cabrera A, Martin A, Barro F (2002) In-situ comparative mapping (ISCM) of Glu-1 loci in Triticum and Hordeum. Chromosome Res 10:49–54. https://doi.org/10.1023/a:1014270227360
Cauderon Y, Rhind JM (1976) Effect on wheat of an Agropyron chromosome carrying stripe rust resistance. Ann Amelior Plantes 26:745–749
Chen Q, Jahier J, Cauderon Y (1989) Production and cytogenetical studies of hybrids between Triticum aestivum L. Thell and Agropyron cristatum (L.) Gaertn. C R Acad Sci Ser III Sci Vie Life Sci 308:425–430
Chen Q, Lu YL, Jahier J, Bernard M (1994) Identification of wheat-Agropyron cristatum monosomic addition lines by RFLP analysis using a set of assigned wheat DNA probes. Theor Appl Genet 89:70–75. https://doi.org/10.1007/BF00226985
Cheng D, Jin-Peng Z, Xiao-Yang WU et al (2012) Development of EST markers specific to Agropyron cristatum chromosome 6P in common wheat background. Acta Agron Sin 38:1791–1801. https://doi.org/10.3724/SP.J.1006.2012.01791
Cherif-Mouaki S, Said M, Alvarez JB, Cabrera A (2011) Sub-arm location of prolamin and EST-SSR loci on chromosome 1H(ch) from Hordeum chilense. Euphytica 178:63–69. https://doi.org/10.1007/s10681-010-0268-y
Choi H-W, Koo D-H, Bang K-H et al (2009) FISH and GISH analysis of the genomic relationships among species. Genes Genom 31:99–105. https://doi.org/10.1007/BF03191143
Consortium (IWGSC) TIWGS, Investigators IR principal, Appels R et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361:eaar7191. https://doi.org/10.1126/science.aar7191
Copete A, Cabrera A (2017) Chromosomal location of genes for resistance to powdery mildew in Agropyron cristatum and mapping of conserved orthologous set molecular markers. Euphytica 213:189. https://doi.org/10.1007/s10681-017-1981-6
Copete A, Moreno R, Cabrera A (2018) Characterization of a world collection of Agropyron cristatum accessions. Genet Resour Crop Evol 65:1455–1469. https://doi.org/10.1007/s10722-018-0630-9
Danilova TV, Friebe B, Gill BS (2012) Single-copy gene fluorescence in situ hybridization and genome analysis: Acc-2 loci mark evolutionary chromosomal rearrangements in wheat. Chromosoma 121:597–611. https://doi.org/10.1007/s00412-012-0384-7
Devos KM, Atkinson MD, Chinoy CN et al (1993) Chromosomal rearrangements in the rye genome relative to that of wheat. Theor Appl Genet 85:673–680. https://doi.org/10.1007/BF00225004
Dewey DR (1960) Salt tolerance of twenty-five strains of Agropyron. Crop Sci 52:631–635. https://doi.org/10.2134/agronj1960.00021962005200110006x
Dewey DR (1962) Breeding crested wheatgrass for salt tolerance. Crop Sci 2:403–407. https://doi.org/10.2135/cropsci1962.0011183X000200050012x
Dewey DR (1984) The genomic system of classification as a guide to intergeneric hybridization with the perennial triticeae. In: Gustafson JP (ed) Gene manipulation in plant improvement. Springer, Boston, pp 209–279
Dewey DR, Asay KH (1982) Cytogenetic and taxonomic relationships among three diploid crested wheatgrasses. Crop Sci 22:645–650. https://doi.org/10.2135/cropsci1982.0011183X002200030052x
Dong YS, Zhou RH, Xu SJ et al (1992) Desirable characteristics in perennial Triticeae collected in China for wheat improvement. Hereditas 116:175–178. https://doi.org/10.1111/j.1601-5223.1992.tb00224.x
Endo TR (2015) Gametocidal genes. In: Molnár-Láng M, Ceoloni C, Doležel J (eds) Alien introgression in wheat: cytogenetics, molecular biology, and genomics. Springer, Cham, pp 121–131
Feuillet C, Langridge P, Waugh R (2008) Cereal breeding takes a walk on the wild side. Trends Genet 24:24–32. https://doi.org/10.1016/j.tig.2007.11.001
Forster BP, Gorham J, Miller TE (1987) Salt tolerance of an amphiploid between Triticum aestivum and Agropyron junceum. Plant Breed 98:1–8. https://doi.org/10.1111/j.1439-0523.1987.tb01083.x
Friebe B, Zeller FJ, Mukai Y et al (1992) Characterization of rust-resistant wheat-Agropyron intermedium derivatives by C-banding, in situ hybridization and isozyme analysis. Theor Appl Genet 83:775–782. https://doi.org/10.1007/BF00226697
Friebe B, Jiang J, Raupp WJ et al (1996) Characterization of wheat-alien translocations conferring resistance to diseases and pests: current status. Euphytica 91:59–87. https://doi.org/10.1007/BF00035277
Fukui K, Kamisugi Y, Sakai F (1994) Physical mapping of 5S rDNA loci by direct-cloned biotinylated probes in barley chromosomes. Genome 37:105–111. https://doi.org/10.1139/g94-013
Gaál E, Valárik M, Molnár I et al (2018) Identification of COS markers specific for Thinopyrum elongatum chromosomes preliminary revealed high level of macrosyntenic relationship between the wheat and T. elongatum genomes. PLoS ONE 13:e0208840. https://doi.org/10.1371/journal.pone.0208840
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885. https://doi.org/10.1093/nar/7.7.1869
Gong W, Gong W, Han R et al (2016) Chromosome arm-specific markers from Aegilops searsii permits targeted introgression. Biologia 71:87–92. https://doi.org/10.1515/biolog-2016-0003
Gong W, Han R, Li H et al (2017) Agronomic traits and molecular marker identification of wheat-Aegilops caudata addition lines. Front Plant Sci 8:1743. https://doi.org/10.3389/fpls.2017.01743
Han F, Fedak G, Guo W, Liu B (2005) Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids. Genetics 170:1239–1245. https://doi.org/10.1534/genetics.104.039263
Han H, Bai L, Su J et al (2014) Genetic rearrangements of six wheat-Agropyron cristatum 6P addition lines revealed by molecular markers. PLoS ONE 9:e91066. https://doi.org/10.1371/journal.pone.0091066
Han H, Liu W, Lu Y et al (2017) Isolation and application of P genome-specific DNA sequences of Agropyron Gaertn. in Triticeae. Planta 245:425–437. https://doi.org/10.1007/s00425-016-2616-1
Howard T, Rejab NA, Griffiths S et al (2011) Identification of a major QTL controlling the content of B-type starch granules in Aegilops. J Exp Bot 62:2217–2228. https://doi.org/10.1093/jxb/erq423
Kato A, Lamb JC, Birchler JA (2004) Chromosome painting using repetitive DNA sequences as probes for somatic chromosome identification in maize. Proc Natl Acad Sci USA 101:13554–13559. https://doi.org/10.1073/pnas.0403659101
Kato A, Albert PS, Vega JM, Birchler JA (2006) Sensitive fluorescence in situ hybridization signal detection in maize using directly labeled probes produced by high concentration DNA polymerase nick translation. Biotech Histochem 81:71–78. https://doi.org/10.1080/10520290600643677
Knott DR (1964) The effect on wheat of an Agropyron chromosome carrying rust resistance. Can J Genet Cytol 6:500–507. https://doi.org/10.1139/g64-064
Knott DR (1968) Translocations involving Triticum chromosomes and Agropyron chromosomes carrying rust resistance. Can J Genet Cytol 10:695–696. https://doi.org/10.1139/g68-087
Le HT, Armstrong KC, Miki B (1989) Detection of rye DNA in wheat-rye hybrids and wheat translocation stocks using total genomic DNA as a probe. Plant Mol Biol Rep 7:150–158. https://doi.org/10.1007/BF02669631
Li L, Li X, Li P et al (1997) Establishment of wheat-Agropyron cristatum alien addition lines. I. Cytology of F_(3), F_(2) BC_(1), BC_(4), and BC_(3) F_(1) progenies. Acta Genet Sin 24:154–159
Li L, Yang X, Zhou R et al (1998) Establishment of wheat-Agropyron cristatum alien addition lines: II. Identification of alien chromosomes and analysis of development approaches. Acta Genet Sin 25:538–544
Li S-F, Zhou H-B, Li L-H et al (2007) Isolation and characterization of high-molecular-weight glutenin subunit genes in Agropyron cristatum. Acta Agron Sin 33:63–69
Li Q, Lu Y, Pan C et al (2016) Characterization of a novel wheat-Agropyron cristatum 2P disomic addition line with powdery mildew resistance. Crop Sci 56:2390–2400. https://doi.org/10.2135/cropsci2015.10.0638
Limin AE, Fowler DB (1987) Cold hardiness of forage grasses grown on the Canadian prairies. Can J Plant Sci 67:1111–1115. https://doi.org/10.4141/cjps87-150
Limin AE, Fowler DB (1990) An interspecific hybrid and amphiploid produced from Triticum aestivum crosses with Agropyron cristatum and Agropyron desertorum. Genome 33:581–584. https://doi.org/10.1139/g90-085
Linc G, Gaál E, Molnár I et al (2017) Molecular cytogenetic (FISH) and genome analysis of diploid wheatgrasses and their phylogenetic relationship. PLoS ONE 12:e0173623. https://doi.org/10.1371/journal.pone.0173623
Littlejohn GM (1988) Salt tolerance of amphiploids and derivatives of crosses between wheat and wild Thinopyrum species. In: 7th Int wheat genet symp, Cambridge, pp 845–849
Liu W, Liu W, Wu J et al (2010) Analysis of genetic diversity in natural populations of Psathyrostachys huashanica Keng using microsatellite (SSR) markers. Agric Sci China 9:463–471. https://doi.org/10.1016/S1671-2927(09)60118-8
Löve Á (1982) Generic evolution of the wheatgrasses. Biol Zbl 101:199–212
Löve A (1984) Conspectus of the Triticeae. Feddes Repert 95:425–521. https://doi.org/10.1002/fedr.4910950702
Lu M, Lu Y, Li H et al (2016) Transferring desirable genes from Agropyron cristatum 7P chromosome into common wheat. PLoS ONE 11:e0159577. https://doi.org/10.1371/journal.pone.0159577
Luan Y, Wang X, Liu W et al (2010) Production and identification of wheat-Agropyron cristatum 6P translocation lines. Planta 232:501–510. https://doi.org/10.1007/s00425-010-1187-9
Ma X-F, Fang P, Gustafson JP (2004) Polyploidization-induced genome variation in triticale. Genome 47:839–848. https://doi.org/10.1139/g04-051
Martín A, Cabrera A, Esteban E et al (1999) A fertile amphiploid between diploid wheat (Triticum tauschii) and crested wheat grass (Agropyron cristatum). Genome 42:519–524. https://doi.org/10.1139/gen-42-3-519
Martín AC, Rey M-D, Shaw P, Moore G (2017) Dual effect of the wheat Ph1 locus on chromosome synapsis and crossover. Chromosoma 126:669–680. https://doi.org/10.1007/s00412-017-0630-0
McGuire PE, Dvořák J (1981) High salt-tolerance potential in wheatgrasses. Crop Sci 21:702–705. https://doi.org/10.2135/cropsci1981.0011183X002100050018x
Miller TE, Reader SM, Chapman V (1982) The addition of Hordeum chilense chromosomes to wheat. In: Broertjes C (ed) Proceedings of international symposium on Eucarpia on induced variability in plant breeding, Wageningen, Pudoc, pp 79–81
Molnár I, Šimková H, Leverington-Waite M et al (2013) Syntenic relationships between the U and M genomes of Aegilops, wheat and the model species Brachypodium and Rice as revealed by COS markers. PLoS ONE 8:e70844. https://doi.org/10.1371/journal.pone.0070844
Molnár I, Kubaláková M, Šimková H et al (2014) Flow cytometric chromosome sorting from diploid progenitors of bread wheat, T. urartu, A. speltoides and A. tauschii. Theor Appl Genet 127:1091–1104. https://doi.org/10.1007/s00122-014-2282-2
Molnár I, Vrána J, Burešová V et al (2016) Dissecting the U, M, S and C genomes of wild relatives of bread wheat (Aegilops spp.) into chromosomes and exploring their synteny with wheat. Plant J 88:452–467. https://doi.org/10.1111/tpj.13266
Molnár-Láng M, Novotny C, Linc G, Naoy ED (2005) Changes in the meiotic pairing behaviour of a winter wheat-winter barley hybrid maintained for a long term in tissue culture, and tracing the barley chromatin in the progeny using GISH and SSR markers. Plant Breed 124:247–252. https://doi.org/10.1111/j.1439-0523.2005.01097.x
Nagy ED, Molnár I, Schneider A et al (2006) Characterization of chromosome-specific S-SAP markers and their use in studying genetic diversity in Aegilops species. Genome 49:289–296. https://doi.org/10.1139/g05-109
Nasuda S, Friebe B, Gill BS (1998) Gametocidal genes induce chromosome breakage in the interphase prior to the first mitotic cell division of the male gametophyte in wheat. Genetics 149:1115–1124
Ochoa V, Madrid E, Said M et al (2015) Molecular and cytogenetic characterization of a common wheat-Agropyron cristatum chromosome translocation conferring resistance to leaf rust. Euphytica 201:89–95. https://doi.org/10.1007/s10681-014-1190-5
Parida SK, Kumar KAR, Dalal V et al (2006) Unigene derived microsatellite markers for the cereal genomes. Theor Appl Genet 112:808–817. https://doi.org/10.1007/s00122-005-0182-1
Perničková K, Koláčková V, Lukaszewski AJ et al (2019) Instability of alien chromosome introgressions in wheat associated with improper positioning in the nucleus. Int J Mol Sci 20:1448. https://doi.org/10.3390/ijms20061448
Quraishi UM, Abrouk M, Bolot S et al (2009) Genomics in cereals: from genome-wide conserved orthologous set (COS) sequences to candidate genes for trait dissection. Funct Integr Genom 9:473–484. https://doi.org/10.1007/s10142-009-0129-8
Rayburn AL, Gill BS (1985) Use of biotin-labeled probes to map specific DNA sequences on wheat chromosomes. J Hered 76:78–81. https://doi.org/10.1093/oxfordjournals.jhered.a110049
Said M, Cabrera A (2009) A physical map of chromosome 4Hch from H. chilense containing SSR, STS and EST-SSR molecular markers. Euphytica 167:253–259. https://doi.org/10.1007/s10681-009-9895-6
Said M, Recio R, Cabrera A (2012) Development and characterisation of structural changes in chromosome 3Hch from Hordeum chilense in common wheat and their use in physical mapping. Euphytica 188:429–440. https://doi.org/10.1007/s10681-012-0712-2
Said M, Hřibová E, Danilova TV et al (2018) The Agropyron cristatum karyotype, chromosome structure and cross-genome homeology as revealed by fluorescence in situ hybridization with tandem repeats and wheat single-gene probes. Theor Appl Genet 131:2213–2227. https://doi.org/10.1007/s00122-018-3148-9
Said M, Kubaláková M, Karafiátová M et al (2019) Dissecting the complex genome of crested wheatgrass by chromosome flow sorting. Plant Genome 12:180096. https://doi.org/10.3835/plantgenome2018.12.0096
Schneider A, Linc G, Molnár I, Molnár-Láng M (2005) Molecular cytogenetic characterization of Aegilops biuncialis and its use for the identification of 5 derived wheat-Aegilops biuncialis disomic addition lines. Genome 48:1070–1082. https://doi.org/10.1139/g05-062
Schulz-Schaeffer J, Allderdice PW, Creel GC (1963) Segmental allopolyploidy in tetraploid and hexaploid Agropyron species of the crested wheatgrass complex (section Agropyron). Crop Sci 3:525–530. https://doi.org/10.2135/cropsci1963.0011183X000300060021x
Schwarzacher T, Heslop-Harrison P (2000) Practical in situ hybridization, 1st edn. Bios Scientific Pub Ltd, Oxford
Schwarzacher T, Leitch AR, Bennett MD, Heslop-Harrison JS (1989) In situ localization of parental genomes in a wide hybrid. Ann Bot 64:315–324. https://doi.org/10.1093/oxfordjournals.aob.a087847
Sharma HC, Gill BS, Uyemoto JK (1984) High levels of resistance in Agropyron species to barley yellow dwarf and wheat streak mosaic viruses. J Phytopathol 110:143–147. https://doi.org/10.1111/j.1439-0434.1984.tb03402.x
Shi F, Liu K-F, Endo TR, Wang D-W (2005) Inducing rye 1R chromosome structural changes in common wheat cv. Chinese spring by the gametocidal chromosome 2C of Aegilops cylindrica. Acta Genet Sin 32:487–494
Shukle RH, Lampe DJ, Lister RM, Foster JE (1987) Aphid feeding behavior: relationship to barley yellow dwarf virus resistance in Agropyron species. Phytopathology 77:725–729. https://doi.org/10.1094/Phyto-77-725
Soliman MH, Cabrera A, Sillero JC, Rubiales D (2007) Genomic constitution and expression of disease resistance in Agropyron cristatum × durum wheat derivatives. Breed Sci 57:17–21. https://doi.org/10.1270/jsbbs.57.17
Song L, Jiang L, Han H et al (2013) Efficient induction of wheat-Agropyron cristatum 6P translocation lines and GISH detection. PLoS ONE 8:e69501. https://doi.org/10.1371/journal.pone.0069501
Szakács E, Molnár-Láng M (2010) Molecular cytogenetic evaluation of chromosome instability in Triticum aestivum–Secale cereale disomic addition lines. J Appl Genet 51:149–152. https://doi.org/10.1007/BF03195723
Taketa S, Kato J, Takeda K (1995) High crossability of wild barley (Hordeum spontaneum C. Koch) with bread wheat and the differential elimination of barley chromosomes in the hybrids. Theor Appl Genet 91:1203–1209. https://doi.org/10.1007/BF00220930
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066. https://doi.org/10.1126/science.277.5329.1063
Taylor RJ, McCoy GA (1973) Proposed origin of tetraploid species of crested wheatgrass based on chromatographic and karyotypic analyses. Am J Bot 60:576–583. https://doi.org/10.2307/2441382
Triebe B, Mukai Y, Dhaliwal HS et al (1991) Identification of alien chromatin specifying resistance to wheat streak mosaic and greenbug in wheat germ plasm by C-banding and in situ hybridization. Theor Appl Genet 81:381–389. https://doi.org/10.1007/BF00228680
Vogel KP, Arumuganathan K, Jensen KB (1999) Nuclear DNA content of perennial grasses of the triticeae. Crop Sci 39:661–667. https://doi.org/10.2135/cropsci1999.0011183X003900020009x
Whelan EDP (1988) Transmission of a chromosome from decaploid Agropyron elongatum that confers resistance to the wheat curl mite in common wheat. Genome 30:293–298. https://doi.org/10.1139/g88-051
Wu J, Yang X, Wang H et al (2006) The introgression of chromosome 6P specifying for increased numbers of florets and kernels from Agropyron cristatum into wheat. Theor Appl Genet 114:13–20. https://doi.org/10.1007/s00122-006-0405-0
Yang C-T, Fan X, Wang X-L et al (2014) Karyotype analysis of Agropyron cristatum (L.) Gaertner. Caryologia 67:234–237. https://doi.org/10.1080/0144235X.2014.974351
Ye X, Lu Y, Liu W et al (2015) The effects of chromosome 6P on fertile tiller number of wheat as revealed in wheat-Agropyron cristatum chromosome 5A/6P translocation lines. Theor Appl Genet 128:797–811. https://doi.org/10.1007/s00122-015-2466-4
Yu J, Wang J, Lin W et al (2005) The genomes of Oryza sativa: a history of duplications. PLoS Biol 3:e38. https://doi.org/10.1371/journal.pbio.0030038
Zhang H, Jia J, Gale MD, Devos KM (1998) Relationships between the chromosomes of Aegilops umbellulata and wheat. Theor Appl Genet 96:69–75. https://doi.org/10.1007/s001220050710
Zhang H, Bian Y, Gou X et al (2013) Intrinsic karyotype stability and gene copy number variations may have laid the foundation for tetraploid wheat formation. PNAS 110:19466–19471. https://doi.org/10.1073/pnas.1319598110
Zhang J, Zhang J, Liu W et al (2015) Introgression of Agropyron cristatum 6P chromosome segment into common wheat for enhanced thousand-grain weight and spike length. Theor Appl Genet 128:1827–1837. https://doi.org/10.1007/s00122-015-2550-9
Zhang Z, Song L, Han H et al (2017) Physical localization of a locus from Agropyron cristatum conferring resistance to stripe rust in common wheat. Int J Mol Sci 18:2403. https://doi.org/10.3390/ijms18112403
Zhao Y, Xie J, Dou Q et al (2017) Diversification of the P genome among Agropyron Gaertn. (Poaceae) species detected by FISH. Comp Cytogenet 11:495–509. https://doi.org/10.3897/CompCytogen.v11i3.13124
Zhou S, Zhang J, Che Y et al (2018) Construction of Agropyron Gaertn. genetic linkage maps using a wheat 660 K SNP array reveals a homeologous relationship with the wheat genome. Plant Biotechnol J 16:818–827. https://doi.org/10.1111/pbi.12831
Acknowledgements
The authors thank Drs Hana Šimková, Miroslav Valárik, Petr Cápal and Zbyněk Milec from the Institute of Experimental Botany, Olomouc, Czech Republic, for their advice and assistance. We are grateful to Dr Joseph Robins, ARS Forage and Range Research Laboratory, USDA, Logan, USA, for providing the plant material. This research was funded by the Grant Agency of the Czech Republic (award 16-16992S), ERDF project ‘Plants as a tool for sustainable global development’ (No. CZ.02.1.01/0.0/0.0/16_019/0000827), the Hungarian National Research, Development and Innovation Office (K116277), the Marie Curie Fellowship Grant award ‘AEGILWHEAT’ (H2020-MSCA-IF-2016-746253), Grants award AGL2014-52445-R and RTI2018-093367-B-I00 from the Spanish State Research Agency (Ministry of Science, Innovation and Universities), co-financed by the European Regional Development Fund (FEDER) from the European Union.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there are no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Said, M., Parada, A.C., Gaál, E. et al. Uncovering homeologous relationships between tetraploid Agropyron cristatum and bread wheat genomes using COS markers. Theor Appl Genet 132, 2881–2898 (2019). https://doi.org/10.1007/s00122-019-03394-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-019-03394-1