Abstract
Non-coding RNA (ncRNA) species, mainly long non-coding RNAs (lncRNAs) and microRNAs (miRNAs) have been currently imputed for lesser or greater involvement in human erythropoiesis. These RNA subsets operate within a complex circuit with other epigenetic components and transcription factors (TF) affecting chromatin remodeling during cell differentiation. Lymphoma/leukemia-related (LRF) TF exerts higher occupancy on DNA CpG rich sites and is implicated in several differentiation cell pathways and erythropoiesis among them and also directs the epigenetic regulation of hemoglobin transversion from fetal (HbF) to adult (HbA) form by intervening in the γ-globin gene repression. We intended to investigate LRF activity in the evolving landscape of cells’ commitment to the erythroid lineage and specifically during HbF to HbA transversion, to qualify this TF as potential repressor of lncRNAs and miRNAs. Transgenic human erythroleukemia cells, overexpressing LRF and further induced to erythropoiesis, were subjected to expression analysis in high LRF occupancy genetic loci-producing lncRNAs. LRF abundance in genetic loci transcribing for studied lncRNAs was determined by ChIP-Seq data analysis. qPCRs were performed to examine lncRNA expression status. Differentially expressed miRNA pre- and post-erythropoiesis induction were assessed by next-generation sequencing (NGS), and their promoter regions were charted. Expression levels of lncRNAs were correlated with DNA methylation status of flanked CpG islands, and contingent co-regulation of hosted miRNAs was considered. LRF-binding sites were overrepresented in LRF overexpressing cell clones during erythropoiesis induction and exerted a significant suppressive effect towards lncRNAs and miRNA collections. Based on present data interpretation, LRF’s multiplied binding capacity across genome is suggested to be transient and associated with higher levels of DNA methylation.
Key messages
-
During erythropoiesis, LRF displays extensive occupancy across genetic loci.
-
LRF significantly represses subsets of lncRNAs and miRNAs during erythropoiesis.
-
Promoter region CpG islands’ methylation levels affect lncRNA expression.
-
MiRNAs embedded within lncRNA loci show differential regulation of expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Regulation of gene expression based on mutual epigenetic mechanisms, mainly DNA methylation levels, differential transcription of ncRNA species, and recruitment of histone modifying and chromatin remodeling systems, is attracting research attention in recent years. Of particular interest is the regulation of ncRNA expression mediated by TFs imitating the classical protein coding genes’ transcriptional regulation mechanisms. Correct tissue-specific and time-defined production of miRNAs and lncRNAs, of the most studied ncRNA varieties, constitutes the endogenous chromatin signaling towards a synchronized gene expression regulation. Among other ncRNA-dependent cellular processes, human erythropoiesis and hemoglobin switch from embryonic to HbF and to HbA during gestation are orchestrated by specific sets of miRNAs, lncRNAs, and co-operation of TFs [1,2,3,4]. Gene expression modifying TFs and miRNAs targeting and affecting TFs’ concentration, as well as lncRNAs with chromatin modulatory activity and “sponge” functions against miRNAs, participate in a sequential process supervising the hemoglobin switches during human fetal ontogeny.
lncRNAs have long been implicated in human hematopoiesis by contributing to the conformation of an accessible chromatin landscape in a tissue and stage specific manner [5]. Cross-acting of lncRNAs with genome and/or DNA binding proteins suggests an essential role on gene’s expression regulation, which has been already documented in general [6, 7] and more strictly in hematopoietic stem cell differentiation [8]. DANCR-lncRNA (Differentiation Antagonizing Non-Protein Coding) promotes erythroid differentiation [4], whereas MONC- (Megakaryocytic Oncogenic Non-Coding RNA) and MIR100HG-lncRNAs reside within cell nucleus and are involved in the regulation of erythro-megakaryocytic development. Their abnormally high expression has been evaluated in acute megakaryoblastic leukemia (AMKL) [9]. Along erythropoiesis pathway, hemoglobin switch form HbF (α2γ2 chains) to HbA (α2β2 chains), which under normal conditions occurs at the latest stage of gestation before birth, has been combined with BGLT3-lncRNA (Beta Globin Locus Transcript 3) expression [2]. Besides, CpG islands abundance lengthwise lncRNAs’ loci are associated with Polycomb Repressive Complex (PRC) recruitment, which spread chromatin repressive histone marks by promoting histone and DNA methylation, thus regulating downstream gene expression with variable potency [10].
MiRNAs have been also reported to affect hemoglobin switch from HbF to HbA and in certain circumstances to alleviate anemia symptoms in β-hemoglobinopathies patients. Research efforts to reactivate HbF expression in adult β-hemoglobinopathies patients are ongoing with the ultimate goal of benefiting patients from anemia symptoms and support their de-addiction from constant blood transfusions. Repressing effects towards BCL11A (B-Cell CLL/Lymphoma 11A), the most fully studied γ-globin repressor, are exerted by miR-15a/16–1, miR-486-3p, and miR-210; therefore, HbF levels are increased with valuable results for the patients [1, 11, 12]. MiRNAs are also identified to affect other modifying TFs of the β-globin locus, such as miR-26b which targets GATA-1 (Globin Transcription Factor 1), with indirect up-regulating effects on HbF expression [13] and miR-223-3p associated with LMO2 (LIM Domain Only 2) factor, leading to down-regulation of HbF levels [14]. Complex networks involving miRNAs, lncRNAs, other nuclear ncRNA species and mRNAs are also in the frontline of research efforts. A network consisting of UCA1- and ZEB1-AS1-lnRNAs with miR-548j, miR-3646, miR-937, and miR-19b-3p is confirmed to regulate HbF [15], while overexpressed NR_001589- and uc002fcj.1-lncRNAs in a high-HbF patient group were significantly correlated by bioinformatics methodology with certain sets of miRNAs and mRNAs [16] exhibiting an integrated chromatin modulatory capacity as a cluster.
A yet unsolved issue is decoding the pathway directing the epigenetic elements to a favorable repertoire of action during healthy state or disorder. Genome architecture and epigenetic landscape support positively or negatively ncRNA transcription, depending on the conditions prevailing across genetic loci coding for miRNA clusters, lncRNAs, and other ncRNA species, not sufficiently studied. lncRNAs are considered mainly products of RNA polymerase II transcription [17], and several TFs have been proved to control lncRNA expression in a tissue-specific and high-selective manner. Deregulation of TFs’ expression and a consequent tissue-imbalanced lncRNA expression are often sufficient for the development of several human disorders and for malignant cell transformation [18]. Further to other identical characteristics with “ordinary” genes, such as splicing and polyadenylation, many genetic loci-transcribing lncRNAs include CpG islands with potential regulatory role in their expression levels. Also, many genetic loci of miRNA genes encompass CpG islands, and DNA methylation status regulates miRNA expression. Expression abnormalities in a cancer-specific fashion associated with DNA methylation dependency are reported in several tumors, such as miR-31 in breast cancer [19].
So far, CxxC zinc finger protein 1 (or CFP1) has been shown to bind CpGs associated with active transcription start sites and enhance gene expression [20]. Also, other TFs or subunits of complexes with enzymatic activity to chromatin, such as MLL1/2 (myeloid/lymphoid factor), KDM2A/B (lysine demethylase), and TET1 (ten-eleven translocation), have common zinc finger domains and are reported to interact with CpG islands in mammals [21]. LRF belongs to an evolutionarily conserved family of TFs with zinc finger domains and has been shown to have binding preference at CG-rich and CpG island containing promoters in mouse [22]. In humans, LRF among a variety of known functions [23], binds at the 5′ end of BGLT3, encoding for a lncRNA spanning the intergenic sequence between γ- and β-globin gene and simultaneously at hypersensitive sites of β-globin Locus Control Region (βLCR), supporting hemoglobin transversion from HbF to HbA [24].
To assess the expression levels of miRNAs and lncRNAs influenced by LRF and implicated in hemoglobin switch from HbF to HbA, we have developed an in vitro system of transgenic human erythroleukemia cell line K562, over-expressing LRF transcription factor. Natural process of hemoglobin switch in humans is under control of the βLCR and other cis and trans regulatory elements, mainly modifying TFs acting to the β-type globin locus [3, 25], with BCL11A and LRF recognized as the most dominant repressors of HbF; however, LRF’s aggregate of functions still remains elusive. K562 erythroleukemia cell line constitutively expresses HbF although emanated from an adult individual and thus is a well-suited model to study epigenetic mechanisms controlling erythroid commitment and hemoglobin switch form γ- to β-globin expression, which can be obtained upon cells’ treatment with hemin/erythropoietin cocktail. The aim of the present study is to investigate transcriptional regulation of lncRNAs and miRNAs attributed to TF LRF implicated in erythropoiesis and further to hemoglobin switch, leading to β-globin gene prominent inter-connection with LCR and thus favoring its expression against γ-globin.
Materials and methods
Cell culture, transfection, and erythroid induction
Human K562 erythroleukemia cells were cultured and transfected with episomal vector encompassing the coding sequence (exons 1 and 2) of ZBTB7A gene encoding for LRF, as previously described. Reporter gene eGFP has been included within the same transcriptional cassette, and long-term expression of the exogenous gene has been confirmed by fluorescence microscopy (LEICA Microsystems, Heerbrugg, Switzerland). LRF/ZBTB7A expression levels were evaluated and were up to 11-fold increased. In parallel, transfections were carried out with the vector backbone. Results obtained by qPCR analyses displayed insignificant differences versus untransfected K562 cells. Treatment of transfected and untransfected K562 cells with 30 μΜ hemin (AppliChem GmbH, Darmstadt, Germany) and 5 ng/mL erythropoietin (EPO) (Cell Signaling) for 3 h resulted to erythroid differentiation, hemoglobinization, and induction of β-globin expression [24] Genomic DNA and total RNA were extracted pre- and post-erythroid induction of K562 transfected and untransfected cell clones. Four different cell conditions were studied: K562 untransfected (UT), K562 UT post-hemin/EPO induction (UT+hemin/EPO), K562 transfected and overexpressing LRF (LRF-OE), and LRF-OE post-hemin/EPO induction (LRF-OE+hemin/EPO).
ChIP-Seq data analysis
LRF-binding sites were identified by analyzing existing K562 UT, UT+hemin/EPO, LRF-OE, and LRF-OE+hemin/EPO cell samples from chromatin immunoprecipitation assays with antibodies against LRF (Gene Expression Omnibus accession ID GSE200135), combined with next-generation sequencing (NGS) of precipitated DNA fragments (ChIP-Seq), previously conducted by Chondrou et al. [24]. Reads were analyzed with FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to assess quality. Minion [26] was also used to perform an exhaustive search for sequencing contaminants before and after applying Trimgalore (https://github.com/FelixKrueger/TrimGalore) to remove contaminants and low quality reads with parameters “-q 20–length 20.” The remaining reads were aligned to the human reference genome (GRCh38) with STAR v2.7.10a [27] using parameters “–alignIntronMax 1–outFilterMultimapNmax 1–alignEndsType EndToEnd–alignSJoverhangMin 9999–alignSJDBoverhangMin 9999.” Picard (Picard Tools. Broad Institute. http://broadinstitute.github.io/picard/) was subsequently applied to minimize the enrichment of PCR duplicates. Reads that were aligned to the mitochondrial genome were excluded from downstream analyses. LRF peaks were identified with MACS2 [28] with parameters “-g hs–call-summits–extsize 300-p 5e-2,” using the IgG/no-IP ChIP-Seq as background. Peaks from non-canonical chromosomes were removed. The analysis resulted in the characterization of 13,796 peaks in UT K562 cells, 40,166 in UT+hemin/EPO, 35,781 in LRF-OE, and 596,274 in LRF-OE+hemin/EPO.
CpG methylation analysis with pyrosequencing
Pyrosequencing CpG assay was utilized to assess DNA methylation status across CpG islands located at 5′ prime ends of lncRNAs’ genetic loci. Genomic DNA was extracted from K562 cultivated cells (UT, UT+hemin/EPO, LRF-OE, and LRF-OE+hemin/EPO) with phenol:chloroform:isoamyl alcohol in 25:24:1 ratio (Sigma-Aldrich), according to standard protocol [29]. 1.5 μg of each isolated genomic DNA was bisulfite converted, PCR amplified, and sequenced with PyroMark® technology (QIAGEN GmbH, Hilden, Germany), according to manufacturer’s instructions and as previously described [30]. PCR and sequencing primers are listed in Supplementary table 1.
Expression profiles of lncRNAs with RT-qPCRs
Total RNA was extracted from K562 cultivated cells (UT, UT+hemin/EPO, LRF-OE and LRF-OE+hemin/EPO) using Trizol Reagent (Invitrogen Life Technologies, Germany) according to the manufacturer’s instructions. One microgram of total RNA was converted to cDNA by Quantitect Reverse transcription kit (QIAGEN GmbH, Hilden, Germany). lncRNA expression was assessed by qPCRs with sequence specific primer sets (Supplementary table 1), designed to support detection of the main transcripts of each lncRNA genetic loci. Reactions were performed in Rotor-Gene Q instrument (QIAGEN GmbH, Hilden, Germany) using the PowerTrack SYBRGreen Master Mix (Applied Biosystems). All reactions were run in triplicates in at least two independent experiments and normalized to GAPDH control gene. Undetectable expression levels were assigned to lncRNAs with Cq values above acceptable limits (above 32–33 PCR cycles). The 2^- ΔΔCt method or Pfaffl equation with PCR efficiency correction was used for data analysis [31, 32].
Next-generation sequencing (NGS) of miRNAs and data analysis
Total RNA including microRNA species were purified from K562 cultivated cells (UT, UT+hemin/EPO, LRF-OE, and LRF-OE+hemin/EPO), and a library consisted of all four RNA samples was created with QIAseq miRNA library kit (QIAGEN GmbH, Hilden, Germany). As an initial step, adapters were ligated to the 3′ and 5′ ends of miRNAs, and subsequently, universal cDNA synthesis with UMI (Unique Molecular Indices) assignment, cDNA cleanup, library amplification, and library cleanup were performed. By this method, the presence of adapter dimers, the major contaminant in sequencing libraries, is eliminated. Downstream RNA sequencing reactions were performed in Illumina ISEQ (Illumina, Inc., US) instrument in single-ended mode with 75 bps and read length ranging from 1–1.5 million per sample, which is considered adequate, due to the short length of mature miRNAs (21–25 bp). The in-house generated miRNA-Seq samples from K562 UT, UT+hemin/EPO, LRF-OE, and LRF-OE+hemin/EPO cell clones were initially processed with custom Python scripts to remove reads that did not include the complete UMI sequence. Due to the distinctive size distribution of miRNA sequences and the FastQC quality assessment results, Trimgalore was used with parameters “-q 30–length 18.” Three alignment rounds were applied to account for the uniqueness of miRNA Biology related to the fact that some miRNA sequences originate from multiple genomic loci. Reads were initially aligned on mature miRNA sequences derived from miRBase v22.1 [33] using STAR with parameters “–PCR and sequencing primers are 1–alignIntronMax 1–alignEndsType EndToEnd.” Subsequently, reads that aligned with a mismatch on multiple miRNAs were re-aligned without allowing any mismatches. Reads that were flagged as unaligned in the two aforementioned alignment steps were re-aligned to the human reference genome (GRCh38) using STAR with parameters “–outFilterMismatchNmax 1–alignIntronMax 1–PCR and sequencing primers are Local.” Gencode v39 [34] and miRBase annotation files were combined with the uniquely mapped reads from the three alignment rounds to count reads overlapping with genes and miRNAs. Reads that aligned on miRNAs originating from multiple genomic regions were counted only once. Read counts were adjusted based on the identified UMI sequences, and genes/miRNAs with zero counts across all cell states were removed from subsequent analyses (Supplementary Table 2).
Identification of cell-state-specific miRNA expression
DEGseq [35] was applied on miRNA read counts to identify differentially expressed miRNAs between LRF-OE+hemin/EPO and UT+hemin/EPO K562 cells, LRF-OE+hemin/EPO and LRF-OE, LRF-OE+hemin/EPO and UT, UT+hemin/EPO and UT, as well as LRF-OE and UT, respectively, with a log fold-change cutoff of 0.5 and an adjusted p value cutoff of 0.05 (Supplementary Table 3).
lncRNA and miRNA gene promoter characterization
The transcription start site (TSS) of miRNA genes were downloaded from DIANA-miRGen v4 [36] repository that contains CAGE-Seq-derived TSSs as calculated by ADAPT-CAGE algorithm [37]. Promoter regions were defined by extending each TSS 1.5 kb upstream and 1 kb downstream. To achieve maximum sensitivity, the entire DIANA-miRGen set of miRNA gene TSSs was used which resulted in the definition of promoters in hundreds of tissues, cell lines, and primary cells. The promoters of differentially expressed miRNAs were intersected with LRF ChIP-Seq peaks derived from matching cell-states to narrow down the list of LRF:miRNA regulatory interactions.lncRNA TSSs were extracted from the Gencode v39 human annotation files, and promoter regions were calculated by extending TSSs 1.5 kb upstream and 1 kb downstream.
Statistical analysis
Statistical analysis was performed with SPSS software version 20.0. One-way ANOVA with Tukey’s multiple comparisons post hoc tests or t-test was applied to compare expression levels between the different groups. p values < 0.05 was considered statistically significant.
Results
Selection of lncRNAs as LRF targets
lncRNAs selected for the present study met specific criteria; preserved LRF occupancy at promoter regions either co-occurring with CpG islands or not and/or functions related to erythropoiesis were already assigned to these molecules. To define LRF:lncRNA interactions upon hemin/EPO treatment, LRF peaks were divided into three groups using BEDTools intersect [38]: peaks unique to hemin/EPO-treated or untreated K562 cells and peaks found in both conditions. Peaks from the hemin/EPO-treated were annotated based on their overlap with lncRNA gene promoters. lncRNA promoters with overlapping LRF peaks, implying LRF occupancy during erythropoiesis induction, were selected for further study (Table 1).
Among selected lncRNAs, HOXA Antisense RNA (HOTAIRM1), Deleted In Lymphocytic Leukemia 1/2 (DLEU1/2), Maternally Expressed 3 (MEG3), Nuclear Paraspeckle Assembly Transcript 1 (NEAT1), Differentiation Antagonizing Non-Protein Coding RNA (DANCR), Plasmacytoma Variant Translocation 1 (PVT1), Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1), and Imprinted Maternally Expressed Transcript H19 are already associated with hematological disorders [39,40,41,42,43,44,45,46,47]. Additionally, long non-coding RNA transcript sequences, derived from ENSG00000225806, ENSG00000236140, ENSG00000236617 (CHFR-DT), ENSG00000248925 (PDCD6-DT), ENSG00000249494 (DMXL1-DT), ENSG00000254821, ENSG00000263923, ENSG00000267338, and ENSG00000273391 genetic loci, which are found in Homo sapiens and annotated by several databases (ENA, LncBook, Ensembl, LNCipedia, Ensembl/GENCODE, GeneCards), were selected for investigation as well. Publications related to these transcripts (when exist) refer to genome-wide surveys or analysis of RNA-seq data; however, any affiliated molecular processes are not yet evident. All above-mentioned lncRNA transcripts were flanked by CpG islands. Sequences and chromosomal locations of CpG islands were tracked from UCSC Genome browser (https://genome-euro.ucsc.edu/cgi-bin/hgGateway) (Table 1).lncRNAs exempted from neighboring CpG islands’ existence, but encompassing LRF peaks during erythropoiesis induction, were also subjected to expression analysis. BGLT3-lncRNA [2] and transcripts derived from ENSG00000271833, ENSG00000249249, and ENSG00000287483 genetic loci were evaluated for the LRF influence within a CpG island-free context.
In total, 20 lncRNAs were studied for both methylation and expression levels. CpG island 116 of HOTARLM1-lncXRNA, due to its long sequence was sequenced with upstream and downstream sets of primers. Also, CpGs 45 and 78 were located upstream of MEG3 loci and were both subjected to methylation analyses.
LFR abundance and binding in methylated CpG islands during erythropoiesis induction
LRF occupancy was more abundant in LRF-OE clones compared with UT (35,781 and 13,796 LRF peaks, respectively), whereas erythropoiesis induction with hemin/EPO treatment raised LRF affinity to DNA and peaks up to 5.7 × 105 across genome (~ 43.2-fold increase compared to UT) (Figs. 1 and 2A). LRF responded to hemin/EPO treatment (within the 3-h induction) by increasing recognition and density of its binding sites across K562 cells’ genome, probably exerting its regulatory influence on primary and secondary gene loci during cell differentiation process towards the erythroid commitment.
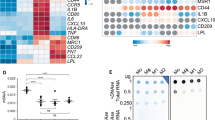
LRF occupancy sites at genetic loci of hypermethylated lncRNAs. MEG 3 (A), PVT1 (B), and ENSG00000225806 (C) displayed methylation levels between 70 and 92% both in UT and LRF-OE K562 clones pre- and post-hemin/EPO induction. LRF-binding sites were widely spread in all aforementioned K562 clones reinforcing the observation that methylation facilitates LRF binding. Methylation of whole region analyzed is presented as mean ± SE. ChIP seq data was retrieved from previous study [24]. CpG islands at 5' end of lncRNAs’ genetic loci are highlighted with orange squares
A LRF/ZBTB7A occupancy sites at lncRNAs genetic loci. Tracks display ChIP-seq data of LRF occupancy in untransfected (UT) and LRF-overexpressing (LRF-OE) K562 cells pre- and post-hemin/EPO induction. Calculated peak enrichment sites are depicted. LRF occupancy was ~ 43.2-fold more abundant in LRF-OE clones after erythropoiesis induction (LRF-OE+hemin/EPO). CpG islands at 5′ end of lncRNAs’ genetic loci are highlighted with orange squares. B Heatmap representing methylation status of CpG islands lying at promoter regions of lncRNAs and displaying medium to low methylation patterns. Methylation elevation is observed only in LRF-OE+hemin/EPO cell subgroup. Each square represents the mean percentage methylation of total sequence analyzed lengthwise each CpG island. A chromatic scale is shown on the right, in which red and green correspond to higher and lower methylation statuses, respectively. C Heatmap depicting expression levels of lncRNAs in UT and LRF-OE K562 cells pre- and post-erythropoiesis induction relative to UT non-induced cells. Cell subgroups of LRF-OE and LRF-OE+hemin/EPO display reduced expression levels of studied lncRNAs. A chromatic scale is shown on the right, in which blue and yellow correspond to a higher and lower expression respectively
CpG islands located 5′ upstream of lncRNA genetic loci were studied with pyrosequencing methylation assay. CpG island 342 of ENSG00000225806-lncRNA, both CpGs 45 and 78 of MEG3-lncRNA, and CpG 94 of PVT1-lncRNA were hypermethylated (methylation levels above 70% and up to 92%) in every single cytosine followed by guanosine deoxynucleotide with methylation potential (Fig. 1). CpG island 35 of ENSG00000236140-lncRNA, CpG island 36 of H19-lncRNA and upstream sequence of CpG 116 of HOTAIRM1-lncRNA displayed medium methylation status (between 50 and 60%) across CpG island’s sequence, whereas downstream sequence of CpG 116 as well as CpGs 175 and 53 of ENSG00000267338- and ENSG00000254821-lncRNAs, respectively, were methylated in low rate, between 16 and 26%. The rest examined lncRNA loci were hypomethylated with methylation mean values below 10% (Table 1). CpG sites with low to medium methylation status, mainly across CpGs 175, 116, 53, 36, and 35, showed high variance in methylation of single sites across the CpG sequence (Supplementary Fig. 1). BGLT3-, ENSG00000287483, ENSG00000249249, and ENSG00000271833 genetic loci lacked CpG islands within 1.5 kb upstream of TSS and were examined only for lncRNAs’ expression levels.
A clear trend for increased methylation levels was obtained in LRF-OE+hemin/EPO cell clones lengthwise every CpG island sequence, whereas cell subgroups of UT, UT+hemin/EPO, and LRF-OE displayed insignificant levels of 5′ methyl-cytosines over unmethylated cytosines in corresponding CpG islands (Fig. 2B). Conclusively, aberrant expression levels of LRF and simultaneous erythropoiesis induction (LRF-OE+hemin/EPO condition) were associated with increased methylation status of CpG islands examined, which facilitated the wide spread of recognition and occupancy by LRF. This result implies the sensing capability and advanced binding of LRF in genomic sites with up-shifted methylation patterns.
lncRNA expression levels correlated to DNA methylation status
Transcription efficacy of studied lncRNA genetic loci demonstrated three differentially expressed groups: those with undetectable expression, those that exhibited equal levels of expression regardless of different cell conditions, and those with decreased expression levels, apparent during LFR-OE and LRF-OE post-erythropoiesis induction. Correlations between DNA methylation status of CpG islands and expression levels led to the following results: transcription was undetectable in ENSG00000225806-, HOTAIRM1-, and MEG3-lncRNAs, derived from highly methylated genetic loci (above 60–70%), and analogous results were obtained from DLEU2-lncRNA and ENSC00000236617-, ENSC00000248925-, ENSC00000249494-, and ENSC00000263923-lncRNAs, comprising the group with hypomethylated CpG islands (bellow 10%) (Fig. 2C). PVT1-lncRNA, which was flanked by a hypermethylated CpG island (methylation levels above 70%) was found exceptionally active. Notably, DLEU2-lncRNA shares the same CpG island with DLEU1-lncRNA, an alternative transcript; however, they were differentially expressed in all experimental conditions of the present study.
Data interpretation of qPCRs particularly focused on cell subgroups of LRF-OE against the UT K562 cells and the LRF-OE with simultaneous induction of erythropoiesis against the corresponding UT subgroup (LRF-OE+hemin/EPO, against UT+hemin/EPO). LRF overexpression (LRF-OE) underscored its effect in DANCR-, BGLT3-, DLEU1-, MALAT1-, NEAT1-, H19-, ENSG00000273391-, ENSG00000236140-, ENSG00000254821-, and ENSG00000287483-lncRNAs, which were downregulated. Transcription levels of DANCR-, BGLT3-, DLEU1-, H19-, ENSG00000273391-, ENSG00000236140-, and ENSG00000287483-lncRNAs were decreased with significant values (p ≤ 0.001–0.05), compared to UT as reference (Fig. 3A). ENSG00000249249-, ENSG00000267338-, ENSG00000271833-, and PVT1-lncRNAs showed stable expression patterns under the same comparisons. These results highlighted the lncRNAs’ expression patterns when LRF is present in high levels within cells.
A, B Relative expression of lncRNAs in LRF overexpressing K562 clones before (LRF-OE) (A) and upon erythropoiesis’ induction (LRF-OE+hemin/EPO) (B). High levels of LRF reported lncRNAs’ expression. Data for BGLT3-lncRNA have been previously [24]. *p < 0.05, **p < 0.01, ***p < 0.001. C Venn diagram presenting common and unique differentially expressed miRNAs in cell sub-conditions. MiRNAs in red cycles are downregulated, those in green cycle are upregulated, and those highlighted in black are either downregulated when LRF was overexpressed or upregulated upon hemin/EPO treatment
In LRF-OE+hemin/EPO cell clones, all mentioned lncRNAs (apart from the non-expressed and DLEU1-lncRNA) were more thoroughly repressed, and further, reduction of expression displayed by DANCR-, BGLT3-, MALAT1-, NEAT1-, ENSG00000273391-, ENSG000002236140-, and ENSG00000287483-lncRNAs was statistically significant compared to UT+hemin/EPO as reference (p ≤ 0.001–0.05) (Fig. 3B). Scores for CpG-free lncRNAs was 50% downregulation (BGLT3- and ENSG00000287483-lncRNAs) and 50% with invariant expression levels (ENSG00000249249- and ENSG00000271833-lncRNAs).
Conclusively, lncRNA expression inhibition in LRF-OE+hemin/EPO subgroup of cells was accompanied by methylation elevation in CpG islands located in lncRNA promoter regions and an extensive spreading of LRF occupancy sites across genomic regions.
Down-regulation of miRNA expression mediated by LRF
Mature miRNAs (21–23 bp in length) differentially expressed between the four cell subgroups (UT, UT+hemin/EPO, LRF-OE, LRF-OE+hemin/EPO) were depicted by NGS and DEGseq [35] data analyses. Two miR clusters including miR182-5p and miR-125a-5p and miR99b-5p and let-7e-5p, respectively, simultaneously with single miRs, miR-486-5p, and miR3529-3p, were significantly upregulated upon erythropoiesis induction in both UT+hemin/EPO and LRF-OE+hemin/EPO, indicating the cell commitment to the erythroid lineage. These results are in consistency with other reported data [48]. MiR-126-5p was a unique case with repressed expression during erythropoiesis induction of UT with hemin/EPO treatment (Supplementary table 3). The top two overexpressed miRs, miR-19a-3p and mir16-5p with scores above 15 × 103 reads (Supplementary table 2), were equally distributed in all cell subgroups.
LRF-OE cell clones exhibited only decreased expression levels for several miRNA sets. The same cluster of miR-125a-5p and miR99b-5p (upregulated during hemin/EPO treatment) was downregulated in LRF overexpressing cells. Also, miR-29a-3p, miR-29c-3p, and miR-10a-5p (member of miR cluster including also miR-182-5p and miR-183) were exclusively downregulated in LRF-OE treated with hemin/EPO. Between LRF-OE versus UT and LRF-OE+hemin/EPO versus UT+hemin/EPO, only downregulated miRNAs with significant values were displayed and are illustrated as Venn diagram (Fig. 3C). Table 2 represents sets of miRNAs downregulated with significant values (adjusted p value < 0.05 and log2FC > │0.5│) and total reads above 100. In consistency with results obtained from lncRNA expression profiles, miRs were systematically suppressed during high levels of LRF expression within transfected cells and some particular miR species prevailed upon erythropoiesis induction (miR-99b-5p and miR-125a-5p), whereas miR29a-3p, miR10a-5p, and miR29c-3p were apparent only in LRF-OE+hemin/EPO. The depicted miRNA sets from NGS followed by bioinformatics analysis are implicated in essential cell pathways presented in the “Discussion” section.
lncRNA and miRNAs embedded in the same genetic loci are independently regulated despite common LRF occupancy
The inclusion of specific miRNAs within the same genetic loci by which the examined lncRNAs are transcribed was further investigated. Also, miRNA promoter regions that are either concurring or located at neighboring regions with lncRNA promoters were determined (Table 3) to map common genetic loci for miRNAs and lncRNAs and assess their potential co-regulation of expression mediated by LRF during erythropoiesis induction.
Promoter regions of miR-136, miR-493, miR-337, and miR-127, hosted within MEG3-lncRNA sequence, are detected in almost all cell subgroups; however, raw read counts of corresponding miRs are ≤ 10, implying insignificance as they are considered as ground noise (Supplementary table 2). Also, MEG3-lncRNA has a hypermethylated promoter and undetectable expression levels in all cell subgroups. Analogous results are displayed by miR-296, miR-10399, and miR-16–1 (with very low reads, ≤ 10) embedded in ENSG00000225806, ENSG00000273391, and DLEU1/2 genetic loci respectively. Although these miRs show insignificant values of expression, the corresponding lncRNA hosts have demonstrated variable (detectable) levels of expression (Figs. 2C and 3A). On the contrary, miR-3613 and miR-15A, derived from DLEU1/2 genetic locus and transcribed towards the same direction with DLEU2-lncRNA, which is silent in all cell subgroups, show high levels of expression. MiR-3613 displays 17–23 and miR-15A with 373–617 read counts among cell subgroups (UT, UT+hemin/EPO, LRF-OE, and LRF-OE+hemin/EPO) (Supplementary table 2).
Moreover, miR-1204, miR-1205, miR-1206, and miR-1207 were not disclosed among differentially expressed or detected miRNA species, whereas they are all located across PVT1-lncRNA genetic locus, found to be activated and expressed. Analogous results were obtained by promoter regions of miR-4449 encompassed within DANCR-lncRNA, miR-612 within NEAT1-lncRNA, and miR-4298 within H19 genetic loci.
Results from lncRNAs’ genetic loci hosting miRNAs or their promoter regions suggest an independent mechanism underlying their expression regulation, although LRF occupancy is apparent across genetic locus, mainly in the LRF-OE+hemin/EPO subgroup. These results confirm certain studies that provide evidence for unconventional function between lncRNAs and encompassed miRNAs hosted in the same genetic loci [49], but further studies are necessary to relate the actual function of lncRNAs to their cognate miRNAs and potential cooperation of TFs regarding regulation of both ncRNA species expression.
Discussion
Eukaryotic cells express selective fractions of the transcriptome, which defines the cell identity and phenotype [50]. Repression of tissue-nonspecific genomic loci is preserved by resident TFs, an effect that can be reversible or more stably down-regulated by general mechanisms of chromatin silencing, phenomena capable to persist through many rounds of cell division and differentiation. TFs’ affinity for DNA-binding sites integrates apart from the intrinsic DNA-binding preferences, interactions with other TFs, co-factors, and chromatin modifiers that are active in simultaneous local and temporal context [51], or affinity may vary depending on methylation patterns in CpG DNA sequences and modifications in histone marks [52].
LRF transcription factor, encoded from the human gene ZBTB7A, has been strongly connected with gene expression control and regulation of lineage fate decisions [53], influencing several branches of hematopoietic differentiation. It mainly represses Bim-mediated apoptosis and binds gene targets of the key erythroid transcription factor GATA1 [54] and also is implicated in γ-globin repression during the final hemoglobin switch to the adult form via a regulatory loop including up-regulation of BGLT3-lncRNA expression [24]. The present work originates from the idea that LRF, apart from protein genes, may also exert a regulatory action towards gene loci of non-coding RNA transcripts. Within this frame, 20 lncRNAs with pronounced LRF occupancy (Table 1) were investigated in terms of expression levels during induction of erythropoiesis (Fig. 3B) and hemoglobin switch in human K562 erythroleukemia cell line. Eight of these lncRNAs have already demonstrated an involvement in hematological disorders: HOTAIRM1 gene is adjacent and antisense to the transcription start site of HOXA1, and its transcription is initiated from the shared promoter segment embedded in a CpG island between the two genes. HOTAIRM1-lncRNA is specifically expressed in the myeloid lineage, most highly in the terminal stage of granulocytic differentiation [39]. DLEU1-lncRNA, DLEU2-lncRNA, and a cluster of neighboring protein-coding tumor suppressor genes, located at 13q14.3, are frequently deregulated in chronic lymphocytic leukemia (CLL) [40]. Notably, miR-15a/16–1, which are embedded within the same locus with DLEU1/2 gene, are expressed with a distinct mode [49] and also confirmed by our analysis. MEG3-lncRNA expression has been suggested as a potential biomarker for therapy response and survival profile in childhood acute lymphoblastic leukemia (cALL) [41], whereas encompasses miRNAs and promoter regions of miRNA species (Table 3). NEAT1-lncRNA is upregulated in multiple myeloma and it is involved in mechanisms of cellular stress response [42]. DANCR-lncRNA, apart from its role to erythroid differentiation [4], positively regulates acute myeloid leukemia chemoresistance to cytarabine by promoting autophagy of AML treated cells and acting as a sponge to decrease miRs abundance that target components of the autophagy machinery [43]. PVT1 maps at 8q24 encodes linear and circular transcript isoforms and encompasses miRNAs (Table 3). PVT1 genomic locus is downstream to MYC, and co-interactions have been described among these two genes. The role of PVT1-lncRNA in hematological malignancies highlights its involvement in disease progression and regulation of immune responses [44]. Malat1-lncRNA depletion in mice (highly conserved sequence with humans) inhibits the erythroid myeloid lymphoid cell proliferation [45]. In humans, MALAT1-lncRNA is recognized as a biomarker for atherosclerosis [46]. Reduced expression of H19 Imprinted Maternally Expressed Transcript has been observed in chronic myeloid leukemia (CML), and myelomonocytic leukemia (CMML), as well as in AML [47], and is also the precursor of miR-675 known to downregulate the retinoblastoma (RB) gene, a fact that multiplies the level of complexity to its functional roles [55]. In the present study, lncRNAs hosting miRNAs or their promoter regions revealed differential expression patterns underlying the distinct regulatory mechanisms guiding their expression.
Additional lncRNA transcripts with apparent LRF peaks covering their promoter regions were also subjected to expression analysis although their roles in chromatin regulation have not been uncovered yet. Seventeen out of 20 lncRNAs examined were flanked by CpG islands either over-methylated and thus preventing transcript expression (apart from PVT1-lncRNA) (Fig. 1) or exhibiting variance in methylation patterns. A trend for increased presence of 5′ methyl-cytosines within CpG islands lying in lncRNA promoter regions was clearly demonstrated by LRF-OE+hemin/EPO cell subgroup (Fig. 2B, Supplementary Fig. 1). lncRNA expression repression was observed mainly in LRF-OE+hemin/EPO subgroup accompanied by increased methylation patterns and a simultaneous intense expansion of the LRF recognition sites lengthwise genomic loci investigated (Figs. 1 and 2A). Across promoters of miRNA species detected to be differentially expressed during LRF-OE and LRF-OE differentiating to the erythroid lineage with hemin/EPO treatment (Table 2), LRF showed increased binding affinity (Supplementary Fig. 2), in consistency with results emanated from lncRNAs. These results suggest the LRF capability to trace secondary binding sites that become visible when methylation levels of DNA are increased.
LRF-binding amplitude shown particularly in cell subgroup LRF-OE+hemin/EPO can be also explained by the availability of alternate direct binding partners. LRF belongs to ZBTB transcription factors family consisting of consensus zinc finger DNA-binding motifs and a BTB domain by which form obligate dimers [56]. Dimerization likely facilitates target DNA binding through the C-terminal zinc finger motifs, but additionally to mediating homodimerization, the BTB dimer forms a favorable scaffold for other ligands. High-throughput assays have revealed cooperative binding interactions at heterodimeric motifs, in some cases completely novel that modify the transcriptional regulation of target genes. The heterodimeric motifs can often be distinct from the individual TF’s in vitro binding preferences and may facilitate binding of TFs to weaker binding motifs [57]. LRF has already demonstrated its action as transcriptional repressor, with histone‐modifying complexes’ recruitment capability, that sequentially package the genetic locus transforming chromatin to non-accessible state [58]. These events prevent genes (including genes encoding for non-protein transcripts) from being transcribed and form the basis of stable epigenetic programs controlling gene loci expression potentially throughout differentiation processes, such as erythropoiesis induction.
As established by miRNA profiling across cell subgroups, many of the upregulated miRNAs expressed only when cells were treated with hemin/EPO to induce cell commitment to the red lineage play characteristic roles during hemopoiesis, such as miR cluster 125a/99b/let-7e and miR-10a and miR-29 family, which promote the hematopoietic stem cell (HSC) self-renewal and also have a prominent attribute to discrete hematopoiesis stages [48, 59]. These sets were all significantly repressed in the LRF overexpressing mode and the phenomenon intensified during terminal differentiation of K562 cells by erythropoiesis induction (Fig. 3B and C).
In cell subgroups of LRF-OE and LRF-OE+hemin/EPO, only significantly downregulated miRNA species were obtained (Table 2). MiR-15b-5p has been reported to function as both oncogene and tumor suppressor in several human cancers, while is sponged by lncRNAs, PVT1- and MALAT1-lncRNAs among them [60]. In the present study, miR-15b-5p was reduced, relevant to lncRNA repressed expression. Deregulation of miR29b-3p, which targets BCL11A transcription factor, is in the same direction with the observation that, during K562 differentiation, the expression of γ-globin decreases (mediated by BCL11A enhancement) with an anti-diametric increase of β-globin expression [11, 24, 61]. MiR-29b, miR-29a, and miR-29c are generated from two primary transcripts and compose the miR-29 family, located in chromosomes 7q32.3 and 1q32.2, respectively. They are highly conserved in mammals, and members of the family are implicated in human disorders such as osteoarthritis, osteoporosis, cardiorenal, and immune disease [62]. MiR-26b-5p, miR-185-5p, and miR-191-5p in peripheral blood mononuclear cells target genes related to the myo-inositol metabolism [63]. Pathway analysis (mirPath v.3, via DIANATOOLS) highlighted various signal transduction pathways that were associated to miR-142-5p, including TGF-β, signaling mediated by MAPK, the insulin/IGF pathway, as well as Erb and mTOR signaling [64]. MiR30e-5p expression is suppressed by IFN-α, a pivotal and abundant pro-inflammatory cytokine, and miR-223-3p has been assigned as a signature of response to anti-TNF-α therapy (TNF: tumor necrosis factor) [65, 66]. MiR-128-3p has been identified to target ZC3H12D, a member of the CCCH-type zinc finger-containing proteins family, which negatively regulates Toll-like receptor signaling and suppresses inflammatory cytokines as well as NF-κB [67, 68]. MiR-93-5p has been associated with inhibition of the epithelial protein lost in neoplasm (EPLIN) expression, a protein involved in regulating actin dynamics and cell mobility, thus has been proposed as novel therapeutic factor for inhibiting cancer angiogenesis and progression [69]. Conclusively, miRNA species found significantly repressed by LRF are implicated in myo-inositol metabolism [63], TGF-β, MAPK, insulin/IGF, Erb, and mTOR signaling pathway [64] expression of inflammatory cytokines [65, 66] and targeting transcription factors essential for several cell functions [67,68,69] with an indirect potential towards the erythropoiesis process.
Conclusions
This study provided clear evidence for the repressing capability of LRF in ncRNA molecules, mainly lncRNAs and sets of miRNAs. lncRNAs exhibited differential methylation and expression patterns during LRF overexpression status and further, during erythropoiesis terminal differentiation with concurrent hemoglobin switch form γ- to β-globin. Also, specific miRNA species were significantly downregulated, as yet, remains to be elucidated the actual role of this network of non-coding RNAs towards the last stage of hemopoiesis and simultaneous transition from fetal to adult stage of hemoglobin production, in human non-malignant tissue-specific cell lines. Gene silencing is not necessarily dependent on the continuous residence of a sequence‐specific repressor at a control element. Assays that detect genome‐wide TF‐binding activity, such as ChIP‐seq typically, provide a static snapshot of occupancy precisely at the time point of the assay; however, LRF potentially exerts transcriptional repression based in DNA tethering by a fleeting way.
Availability of data and material
Data generated in the present study is included in supplementary material.
Abbreviations
- AMKL :
-
Acute megakaryoblastic leukemia
- AML :
-
Acute myeloid leukemia
- BCL11A :
-
B-cell CLL/Lymphoma 11A
- BGLT3 :
-
Beta Globin Locus Transcript 3
- cALL :
-
Childhood Acute lymphoblastic leukemia
- CFP1 :
-
CxxC zinc finger protein 1
- CHFR-DT :
-
CHFR divergent transcript
- ChIP-Seq :
-
Chromatin immunoprecipitation sequencing
- CLL :
-
Chronic lymphocytic leukemia
- CML :
-
Chronic myeloid leukemia
- CMML :
-
Chronic myelomonocytic leukemia
- DANCR :
-
Differentiation Antagonizing Non-Protein Coding RNA
- DLEU1/2:
-
Deleted in Lymphocytic Leukemia 1/2
- DMXL1-DT :
-
DMXL1 divergent transcript
- eGFP :
-
Enhanced green fluorescent protein
- EPLIN :
-
Epithelial protein lost in neoplasm
- EPO :
-
Erythropoietin
- GAPDH :
-
Glyceraldehyde-3-phosphate dehydrogenase
- GATA-1 :
-
Globin Transcription Factor 1
- H19 :
-
Imprinted Maternally Expressed Transcript H19
- HbA :
-
Adult hemoglobin (hemoglobin A)
- HbF :
-
Fetal hemoglobin (hemoglobin F)
- HOTAIRM1 :
-
HOXA antisense RNA
- HSC :
-
Hematopoietic stem cell
- IFN-α :
-
Interferon alpha
- IGF :
-
Insulin-like growth factor
- KDM2A/B :
-
Lysine-specific demethylase
- LMO2 :
-
LIM Domain Only 2
- lncRNA :
-
Long non-coding RNA
- LRF :
-
Leukemia-related factor
- LRF-OE :
-
K562 transfected and overexpressing LRF
- LRF-OE + hemin/EPO :
-
LRF overexpressing post hemin/EPO induction
- MALAT1 :
-
Metastasis-Associated Lung Adenocarcinoma Transcript 1
- MAPK :
-
Mitogen-activated protein kinase
- MEG3 :
-
Maternally expressed 3
- miRNA :
-
Micro RNA
- MLL1/2 :
-
Myeloid/lymphoid factor
- MONC :
-
Megakaryocytic oncogenis non-coding
- mRNA :
-
Messenger RNA
- mTOR :
-
Mammalian target of rapamycin
- ncRNA :
-
Non-coding RNA
- NEAT1 :
-
Nuclear Paraspeckle Assembly Transcript 1
- NF-κB :
-
Nuclear factor kappa B
- NGS :
-
Next-generation sequencing
- PDCD6-DT :
-
PDCD6 divergent transcript
- PRC :
-
Polycomb repressive complex
- PVT1 :
-
Plasmacytoma Variant Translocation 1
- RB :
-
Retinoblastoma
- TET1 :
-
Ten-eleven translocation
- TF :
-
Translation factor
- TGF-β :
-
Transforming growth factor beta
- TNF :
-
Tumor necrosis factor
- TSS :
-
Transcription start site
- UCA1 :
-
Urothelial cancer associated 1
- UMI :
-
Unique Molecular Indices
- UT :
-
K562 untransfected
- UT+hemin/EPO :
-
K562 untransfected post hemin/EPO induction
- ZBTB7A :
-
Zinc finger and BTB domain-containing 7A
- ZC3H12D :
-
Zinc finger CCCH-type containing 12D
- ZEB1-AS1 :
-
ZEB1 antisense 1
- βLCR :
-
β-Globin locus control region
References
Lulli V, Romania P, Morsilli O, Cianciulli P, Gabbianelli M, Testa U, Giuliani A, Marziali G et al (2013) MicroRNA-486-3p regulates gamma-globin expression in human erythroid cells by directly modulating BCL11A. PLoS ONE 8(4):e60436
Ivaldi MS, Diaz LF, Chakalova L, Lee J, Krivega I, Dean A et al (2018) Fetal gamma-globin genes are regulated by the BGLT3 long noncoding RNA locus. Blood 132(18):1963–1973
Liu N, Xu S, Yao Q, Zhu Q, Kai Y, Hsu JY, Sakon P, Pinello L, Yuan GC, Bauer DE et al (2021) Transcription factor competition at the gamma-globin promoters controls hemoglobin switching. Nat Genet 53(4):511–520
Ren Y, Zhu J, Han Y, Li P, Wu J, Qu H, Zhang Z, Fang X et al (2021) Regulatory association of long noncoding RNAs and chromatin accessibility facilitates erythroid differentiation. Blood Adv 5(23):5396–5409
Paralkar VR, Weiss MJ (2013) Long noncoding RNAs in biology and hematopoiesis. Blood 121(24):4842–4846
Sun Q, Hao Q, Prasanth KV (2018) Nuclear long noncoding RNAs: key regulators of gene expression. Trends in genetics : TIG 34(2):142–157
Luo S, Lu JY, Liu L, Yin Y, Chen C, Han X, Wu B, Xu R, Liu W, Yan P et al (2016) Divergent lncRNAs regulate gene expression and lineage differentiation in pluripotent cells. Cell Stem Cell 18(5):637–652
Delas MJ, Jackson BT, Kovacevic T, Vangelisti S, Munera Maravilla E, Wild SA, Stork EM, Erard N, Knott SRV, Hannon GJ et al (2019) lncRNA Spehd regulates hematopoietic stem and progenitor cells and is required for multilineage differentiation. Cell Rep 27(3):719–729 e716
Emmrich S, Streltsov A, Schmidt F, Thangapandi VR, Reinhardt D, Klusmann JH (2014) LincRNAs MONC and MIR100HG act as oncogenes in acute megakaryoblastic leukemia. Mol Cancer 13:171
Schertzer MD, Braceros KCA, Starmer J, Cherney RE, Lee DM, Salazar G, Justice M, Bischoff SR, Cowley DO, Ariel P et al (2019) lncRNA-induced spread of polycomb controlled by genome architecture, RNA abundance, and CpG island DNA. Mol Cell 75(3):523–537 e510
Sankaran VG, Menne TF, Xu J, Akie TE, Lettre G, Van Handel B, Mikkola HK, Hirschhorn JN, Cantor AB, Orkin SH et al (2008) Human fetal hemoglobin expression is regulated by the developmental stage-specific repressor BCL11A. Science 322(5909):1839–1842
Gasparello J, Fabbri E, Bianchi N, Breveglieri G, Zuccato C, Borgatti M, Gambari R, Finotti A et al (2017) BCL11A mRNA Targeting by miR-210: a possible network regulating gamma-globin gene expression. Int J Mol Sci 18(12)
Alijani S, Alizadeh S, Kazemi A, Khatib ZK, Soleimani M, Rezvani M, Minayi N, Karami F, Tayebi B et al (2014) Evaluation of the effect of miR-26b Up-regulation on HbF expression in erythroleukemic K-562 cell line. Avicenna J Med Biotechnol 6(1):53–56
Sun KT, Huang YN, Palanisamy K, Chang SS, Wang IK, Wu KH, Chen P, Peng CT, Li CY et al (2017) Reciprocal regulation of gamma-globin expression by exo-miRNAs: relevance to gamma-globin silencing in beta-thalassemia major. Sci Rep 7(1):202
Rahaman M, Bhowmick C, Komanapalli J, Mukherjee M, Byram PK, Shukla PC, Dolai TK, Chakravorty N et al (2021) Debunking the “junk”: unraveling the role of lncRNA–miRNA–mRNA networks in fetal hemoglobin regulation. BioRxiv
Jia W, Jia S, Chen P, He Y et al (2019) Construction and analysis of a long non-coding RNA (lncRNA)-associated ceRNA network in beta-thalassemia and hereditary persistence of fetal hemoglobin. Medical Science Monitor: International Medical Journal of Experimental and Clinical Research 25:7079–7086
Ponting CP, Oliver PL, Reik W et al (2009) Evolution and unctions of long noncoding RNAs. Cell 136(4):629–641
Guo Q, Wang J, Gao Y, Li X, Hao Y, Ning S, Wang P et al (2020) Dynamic TF-lncRNA regulatory networks revealed prognostic signatures in the development of ovarian cancer. Front Bioeng Biotechnol 8:460
Augoff K, McCue B, Plow EF, Sossey-Alaoui K et al (2012) miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer 11:5
van de Lagemaat LN, Flenley M, Lynch MD, Garrick D, Tomlinson SR, Kranc KR, Vernimmen D et al (2018) CpG binding protein (CFP1) occupies open chromatin regions of active genes, including enhancers and non-CpG islands. Epigenetics Chromatin 11(1):59
Long HK, Blackledge NP, Klose RJ et al (2013) ZF-CxxC domain-containing proteins, CpG islands and the chromatin connection. Biochem Soc Trans 41(3):727–740
Ramos Pittol JM, Oruba A, Mittler G, Saccani S et al (2018) D vE: Zbtb7a is a transducer for the control of promoter accessibility by NF-kappa B and multiple other transcription factors. PLoS Biol 16(5):e2004526
Constantinou C, Spella M, Chondrou V, Patrinos GP, Papachatzopoulou A, Sgourou A et al (2019) The multi-faceted functioning portrait of LRF/ZBTB7A. Hum Genomics 13(1):66
Chondrou V, Shaukat AN, Psarias G, Athanasopoulou K, Iliopoulou E, Damanaki A, Stathopoulos C, Sgourou A et al (2022) LRF promotes indirectly advantageous chromatin conformation via BGLT3-lncRNA expression and switch from fetal to adult hemoglobin. Int J Mol Sci 23(13)
Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P et al (2000) Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell 5(2):377–386
Davis MP, van Dongen S, Abreu-Goodger C, Bartonicek N, Enright AJ et al (2013) Kraken: a set of tools for quality control and analysis of high-throughput sequence data. Methods 63(1):41–49
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR et al (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W et al (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9(9):R137
Albarino CG, Romanowski V (1994) Phenol extraction revisited: a rapid method for the isolation and preservation of human genomic DNA from whole blood. Mol Cell Probes 8(5):423–427
Vlaikou AM, Kouroupis D, Sgourou A, Markopoulos GS, Bagli E, Markou M, Papadopoulou Z, Fotsis T, Nakos G, Lekka ME et al (2017) Mechanical stress affects methylation pattern of GNAS isoforms and osteogenic differentiation of hAT-MSCs. Biochim Biophys Acta 1864(8):1371–1381
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25(4):402–408
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29(9):e45
Kozomara A, Birgaoanu M, Griffiths-Jones S et al (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47(D1):D155–D162
Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S et al (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22(9):1760–1774
Wang L, Feng Z, Wang X, Wang X, Zhang X et al (2010) DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26(1):136–138
Perdikopanis N, Georgakilas GK, Grigoriadis D, Pierros V, Kavakiotis I, Alexiou P, Hatzigeorgiou A et al (2021) DIANA-miRGen v4: indexing promoters and regulators for more than 1500 microRNAs. Nucleic Acids Res 49(D1):D151–D159
Georgakilas GK, Perdikopanis N, Hatzigeorgiou A et al (2020) Solving the transcription start site identification problem with ADAPT-CAGE: a Machine Learning algorithm for the analysis of CAGE data. Sci Rep 10(1):877
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842
Zhang X, Weissman SM, Newburger PE et al (2014) Long intergenic non-coding RNA HOTAIRM1 regulates cell cycle progression during myeloid maturation in NB4 human promyelocytic leukemia cells. RNA Biol 11(6):777–787
Garding A, Bhattacharya N, Claus R, Ruppel M, Tschuch C, Filarsky K, Idler I, Zucknick M, Caudron-Herger M, Oakes C et al (2013) Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the in cis downregulation of a gene cluster that targets NF-kB. PLoS Genet 9(4):e1003373
Gao W (2021) Long non-coding RNA MEG3 as a candidate prognostic factor for induction therapy response and survival profile in childhood acute lymphoblastic leukemia patients. Scand J Clin Lab Invest 81(3):194–200
Taiana E, Ronchetti D, Favasuli V, Todoerti K, Manzoni M, Amodio N, Tassone P, Agnelli L, Neri A et al (2019) Long non-coding RNA NEAT1 shows high expression unrelated to molecular features and clinical outcome in multiple myeloma. Haematologica 104(2):e72–e76
Zhang H, Liu L, Chen L, Liu H, Ren S, Tao Y et al (2021) Long noncoding RNA DANCR confers cytarabine resistance in acute myeloid leukemia by activating autophagy via the miR-874-3P/ATG16L1 axis. Mol Oncol 15(4):1203–1216
Ghetti M, Vannini I, Storlazzi CT, Martinelli G, Simonetti G et al (2020) Linear and circular PVT1 in hematological malignancies and immune response: two faces of the same coin. Mol Cancer 19(1):69
Ma XY, Wang JH, Wang JL, Ma CX, Wang XC, Liu FS et al (2015) Malat1 as an evolutionarily conserved lncRNA, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genomics 16(1):676
Liu L, Tan L, Yao J, Yang L et al (2020) Long non-coding RNA MALAT1 regulates cholesterol accumulation in ox-LDL-induced macrophages via the microRNA-17-5p/ABCA1 axis. Mol Med Rep 21(4):1761–1770
Bock O, Schlue J, Kreipe H et al (2003) Reduced expression of H19 in bone marrow cells from chronic myeloproliferative disorders. Leukemia 17(4):815–816
Nath A, Rayabaram J, Ijee S, Bagchi A, Chaudhury AD, Roy D, Chambayil K, Singh J, Nakamura Y, Velayudhan SR et al (2021) Comprehensive analysis of microRNAs in human adult erythropoiesis. Cells 10(11)
Morenos L, Chatterton Z, Ng JL, Halemba MS, Parkinson-Bates M, Mechinaud F, Elwood N, Saffery R, Wong NC et al (2014) Hypermethylation and down-regulation of DLEU2 in paediatric acute myeloid leukaemia independent of embedded tumour suppressor miR-15a/16-1. Mol Cancer 13:123
Ramskold D, Wang ET, Burge CB, Sandberg R et al (2009) An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5(12):e1000598
Siggers T, Gordan R (2014) Protein-DNA binding: complexities and multi-protein codes. Nucleic Acids Res 42(4):2099–2111
Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, Das PK, Kivioja T, Dave K, Zhong F et al (2017) Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science 356(6337)
Lunardi A, Guarnerio J, Wang G, Maeda T, Pandolfi PP et al (2013) Role of LRF/Pokemon in lineage fate decisions. Blood 121(15):2845–2853
Maeda T, Ito K, Merghoub T, Poliseno L, Hobbs RM, Wang G, Dong L, Maeda M, Dore LC, Zelent A et al (2009) LRF is an essential downstream target of GATA1 in erythroid development and regulates BIM-dependent apoptosis. Dev Cell 17(4):527–540
Cai X, Cullen BR (2007) The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 13(3):313–316
Stogios PJ, Chen L, Prive GG et al (2007) Crystal structure of the BTB domain from the LRF/ZBTB7 transcriptional regulator. Protein science : a publication of the Protein Society 16(2):336–342
Jolma A, Yin Y, Nitta KR, Dave K, Popov A, Taipale M, Enge M, Kivioja T, Morgunova E, Taipale J et al (2015) DNA-dependent formation of transcription factor pairs alters their binding specificity. Nature 527(7578):384–388
Masuda T, Wang X, Maeda M, Canver MC, Sher F, Funnell AP, Fisher C, Suciu M, Martyn GE, Norton LJ et al (2016) Transcription factors LRF and BCL11A independently repress expression of fetal hemoglobin. Science 351(6270):285–289
Gerrits A, Walasek MA, Olthof S, Weersing E, Ritsema M, Zwart E, van Os R, Bystrykh LV, de Haan G et al (2012) Genetic screen identifies microRNA cluster 99b/let-7e/125a as a regulator of primitive hematopoietic cells. Blood 119(2):377–387
Ghafouri-Fard S, Khoshbakht T, Hussen BM, Jamal HH, Taheri M, Hajiesmaeili M et al (2022) A comprehensive review on function of miR-15b-5p in Malignant and non-malignant disorders. Front Oncol 12:870996
Marttila S, Rovio S, Mishra PP, Seppala I, Lyytikainen LP, Juonala M, Waldenberger M, Oksala N, Ala-Korpela M, Harville E et al (2021) Adulthood blood levels of hsa-miR-29b-3p associate with preterm birth and adult metabolic and cognitive health. Sci Rep 11(1):9203
Horita M, Farquharson C, Stephen LA et al (2021) The role of miR-29 family in disease. J Cell Biochem 122(7):696–715
Sanchez-Mora C, Soler Artigas M, Garcia-Martinez I, Pagerols M, Rovira P, Richarte V, Corrales M, Fadeuilhe C, Padilla N, de la Cruz X et al (2019) Epigenetic signature for attention-deficit/hyperactivity disorder: identification of miR-26b-5p, miR-185-5p, and miR-191-5p as potential biomarkers in peripheral blood mononuclear cells. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology 44(5):890–897
Carini G, Mingardi J, Bolzetta F, Cester A, Bolner A, Nordera G, La Via L, Ieraci A, Russo I, Maggi S et al (2022) miRNome profiling detects miR-101–3p and miR-142–5p as putative blood biomarkers of frailty syndrome. Genes 13(2)
Kim BS, Jung JY, Jeon JY, Kim HA, Suh CH et al (2016) Circulating hsa-miR-30e-5p, hsa-miR-92a-3p, and hsa-miR-223-3p may be novel biomarkers in systemic lupus erythematosus. Hla 88(4):187–193
Castro-Villegas C, Perez-Sanchez C, Escudero A, Filipescu I, Verdu M, Ruiz-Limon P, Aguirre MA, Jimenez-Gomez Y, Font P, Rodriguez-Ariza A et al (2015) Circulating miRNAs as potential biomarkers of therapy effectiveness in rheumatoid arthritis patients treated with anti-TNFalpha. Arthritis Res Ther 17(1):49
Zhu M, Wu Y, Wang Z, Lin M, Su B, Li C, Liang F, Chen X et al (2021) miR-128-3p serves as an oncogenic microRNA in osteosarcoma cells by downregulating ZC3H12D. Oncol Lett 21(2):152
Zhang H, Wang WC, Chen JK, Zhou L, Wang M, Wang ZD, Yang B, Xia YM, Lei S, Fu EQ et al (2015) ZC3H12D attenuated inflammation responses by reducing mRNA stability of proinflammatory genes. Mol Immunol 67(2 Pt B):206–212
Liang L, Zhao L, Zan Y, Zhu Q, Ren J, Zhao X et al (2017) MiR-93-5p enhances growth and angiogenesis capacity of HUVECs by down-regulating EPLIN. Oncotarget 8(63):107033–107043
Funding
Open access funding provided by HEAL-Link Greece. Conducted research and Katerina Athanasopoulou were financially supported by the 80250 (ELKE_EAP) grant of the Hellenic Open University.
Author information
Authors and Affiliations
Contributions
AS conceived and designed study; AK, VC, and GP performed experiments; PX, AK, GKG, YV, and VC interpreted results; AS and GKG drafted the manuscript; AS, YV, and GKG finalized and reviewed. All authors have given approval to the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consent to the publication of the manuscript.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Katerina Athanasopoulou and Vasiliki Chondrou contributed equally to this work.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Athanasopoulou, K., Chondrou, V., Xiropotamos, P. et al. Transcriptional repression of lncRNA and miRNA subsets mediated by LRF during erythropoiesis. J Mol Med 101, 1097–1112 (2023). https://doi.org/10.1007/s00109-023-02352-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-023-02352-1