Abstract
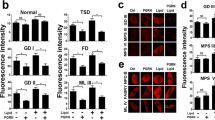
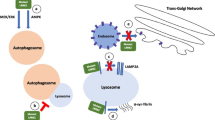
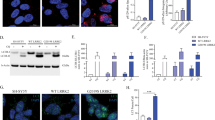
Progranulin (PGRN) is a key regulator of lysosomes, and its deficiency has been linked to various lysosomal storage diseases (LSDs), including Gaucher disease (GD), one of the most common LSD. Here, we report that PGRN plays a previously unrecognized role in autophagy within the context of GD. PGRN deficiency is associated with the accumulation of LC3-II and p62 in autophagosomes of GD animal model and patient fibroblasts, resulting from the impaired fusion of autophagosomes and lysosomes. PGRN physically interacted with Rab2, a critical molecule in autophagosome-lysosome fusion. Additionally, a fragment of PGRN containing the Grn E domain was required and sufficient for binding to Rab2. Furthermore, this fragment significantly ameliorated PGRN deficiency–associated impairment of autophagosome-lysosome fusion and autophagic flux. These findings not only demonstrate that PGRN is a crucial mediator of autophagosome-lysosome fusion but also provide new evidence indicating PGRN’s candidacy as a molecular target for modulating autophagy in GD and other LSDs in general.
Key messages
-
PGRN acts as a crucial factor involved in autophagosome-lysosome fusion in GD.
-
PGRN physically interacts with Rab2, a molecule in autophagosome-lysosome fusion.
-
A 15-kDa C-terminal fragment of PGRN is required and sufficient for binding to Rab2.
-
This PGRN derivative ameliorates PGRN deficiency–associated impairment of autophagy.
-
This study provides new insights into autophagy and may develop novel therapy for GD.






Similar content being viewed by others
Data transparency
All authors confirm that all data and material support their published claims and comply with field standards.
References
Glick D, Barth S, Macleod KF (2010) Autophagy: cellular and molecular mechanisms. J Pathol 221(1):3–12
Settembre C et al (2008) Lysosomal storage diseases as disorders of autophagy. Autophagy 4(1):113–114
Kinghorn KJ, Asghari AM, Castillo-Quan JI (2017) The emerging role of autophagic-lysosomal dysfunction in Gaucher disease and Parkinson’s disease. Neural Regen Res 12(3):380–384
Seranova E et al (2017) Dysregulation of autophagy as a common mechanism in lysosomal storage diseases. Essays Biochem 61(6):733–749
Platt FM (2014) Sphingolipid lysosomal storage disorders. Nature 510(7503):68–75
Chen Y et al (2018) Molecular regulations and therapeutic targets of Gaucher disease. Cytokine Growth Factor Rev 41:65–74
Grabowski GA (2012) Gaucher disease and other storage disorders. Hematology Am Soc Hematol Educ Program 2012:13–18
Pitcairn C, Wani WY, Mazzulli JR (2019) Dysregulation of the autophagic-lysosomal pathway in Gaucher and Parkinson’s disease. Neurobiol Dis 122:72–82
Sun Y, Grabowski GA (2010) Impaired autophagosomes and lysosomes in neuronopathic Gaucher disease. Autophagy 6(5):648–649
Li H et al (2019) Mitochondrial dysfunction and mitophagy defect triggered by heterozygous GBA mutations. Autophagy 15(1):113–130
Moren C et al (2019) GBA mutation promotes early mitochondrial dysfunction in 3D neurosphere models. Aging (Albany NY) 11(22):10338–10355
Li SS et al (2019) Reduction of PGRN increased fibrosis during skin wound healing in mice. Histol Histopathol 34(7):765–774
Jian J, Konopka J, Liu C (2013) Insights into the role of progranulin in immunity, infection, and inflammation. J Leukoc Biol 93(2):199–208
Jian J et al (2016) Progranulin recruits HSP70 to beta-glucocerebrosidase and is therapeutic against Gaucher disease. EBioMedicine 13:212–224
Jian J et al (2018) Progranulin: a key player in autoimmune diseases. Cytokine 101:48–55
Bateman A, Bennett HP (2009) The granulin gene family: from cancer to dementia. BioEssays 31(11):1245–1254
Cui Y, Hettinghouse A, Liu CJ (2019) Progranulin: a conductor of receptors orchestra, a chaperone of lysosomal enzymes and a therapeutic target for multiple diseases. Cytokine Growth Factor Rev 45:53–64
Bateman A, Cheung ST, Bennett HPJ (2018) A brief overview of progranulin in health and disease. Methods Mol Biol 1806:3–15
Baker M et al (2006) Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature 442(7105):916–919
Arrant AE et al (2017) Restoring neuronal progranulin reverses deficits in a mouse model of frontotemporal dementia. Brain 140(5):1447–1465
Gotzl JK et al (2014) Common pathobiochemical hallmarks of progranulin-associated frontotemporal lobar degeneration and neuronal ceroid lipofuscinosis. Acta Neuropathol 127(6):845–860
Mendsaikhan A, Tooyama I, Walker DG (2019) Microglial progranulin: involvement in Alzheimer’s disease and neurodegenerative diseases. Cells 8(3)
Chitramuthu BP, Bennett HPJ, Bateman A (2017) Progranulin: a new avenue towards the understanding and treatment of neurodegenerative disease. Brain 140(12):3081–3104
Tang W et al (2011) The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science 332(6028):478–484
Jian J et al (2013) Progranulin directly binds to the CRD2 and CRD3 of TNFR extracellular domains. FEBS Lett 587(21):3428–3436
Liu C et al (2014) Progranulin-derived Atsttrin directly binds to TNFRSF25 (DR3) and inhibits TNF-like ligand 1A (TL1A) activity. PLoS One 9(3): p. e92743
Li M et al (2014) Progranulin is required for proper ER stress response and inhibits ER stress-mediated apoptosis through TNFR2. Cell Signal 26(7):1539–1548
Chang MC et al (2017) Progranulin deficiency causes impairment of autophagy and TDP-43 accumulation. J Exp Med 214(9):2611–2628
Guo Q et al (2017) Progranulin causes adipose insulin resistance via increased autophagy resulting from activated oxidative stress and endoplasmic reticulum stress. Lipids Health Dis 16(1):25
Zhou B et al (2015) Progranulin induces adipose insulin resistance and autophagic imbalance via TNFR1 in mice. J Mol Endocrinol 55(3):231–243
Liu J et al (2015) PGRN induces impaired insulin sensitivity and defective autophagy in hepatic insulin resistance. Mol Endocrinol 29(4):528–541
Jian J et al (2016) Association between progranulin and Gaucher disease. EBioMedicine 11:127–137
Chen Y et al (2018) Progranulin associates with hexosaminidase A and ameliorates GM2 ganglioside accumulation and lysosomal storage in Tay-Sachs disease. J Mol Med (Berl) 96(12):1359–1373
Elia LP et al (2019) Genetic regulation of neuronal progranulin reveals a critical role for the autophagy-lysosome pathway. J Neurosci 39(17):3332–3344
Coffey EE et al (2014) Lysosomal alkalization and dysfunction in human fibroblasts with the Alzheimer’s disease-linked presenilin 1 A246E mutation can be reversed with cAMP. Neuroscience 263:111–124
K Castillo et al (2013) Measurement of autophagy flux in the nervous system in vivo. Cell Death and Disease 4: p. e917
Shi H et al (2017) Na+/H+ exchanger regulates amino acid-mediated autophagy in intestinal epithelial cells. Cell Physiol Biochem 42:2418–2429
Tooze SA, Yoshimori T (2010) The origin of the autophagosomal membrane. Nat Cell Biol 12(9):831–835
Parzych KR, Klionsky DJ (2014) An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal 20(3):460–473
Mizushima N (2020) The ATG conjugation systems in autophagy. Curr Opin Cell Biol 63:1–10
Romanov J et al (2012) Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J 31(22):4304–4317
Chen M et al (2016) TRIM14 inhibits cGAS degradation mediated by selective autophagy receptor p62 to promote innate immune responses. Mol Cell 64(1):105–119
Lamark T, Svenning S, Johansen T (2017) Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem 61(6):609–624
Danieli A, Martens S (2018) p62-mediated phase separation at the intersection of the ubiquitin-proteasome system and autophagy. J Cell Sci 131(19)
Yoshii SR, Mizushima N (2017) Monitoring and measuring autophagy. Int J Mol Sci 18(9)
Eskelinen EL (2005) Maturation of autophagic vacuoles in mammalian cells. Autophagy 1(1):1–10
Settembre C et al (2008) A block of autophagy in lysosomal storage disorders. Hum Mol Genet 17(1):119–129
Matus S, Valenzuela V, Hetz C (2014) A new method to measure autophagy flux in the nervous system. Autophagy 10(4):710–714
von Muhlinen N (2019) Methods to measure autophagy in cancer metabolism. Methods Mol Biol 1928:149–173
Tanaka Y et al (2017) Progranulin regulates lysosomal function and biogenesis through acidification of lysosomes. Hum Mol Genet 26(5):969–988
Yu L, Chen Y, Tooze SA (2018) Autophagy pathway: cellular and molecular mechanisms. Autophagy 14(2):207–215
Chen Y, Yu L (2018) Development of research into autophagic lysosome reformation. Mol Cells 41(1):45–49
Itakura E, Kishi-Itakura C, Mizushima N (2012) The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell 151(6):1256–1269
Takats S et al (2018) Small GTPases controlling autophagy-related membrane traffic in yeast and metazoans. Small GTPases 9(6):465–471
Nakamura S, Yoshimori T (2017) New insights into autophagosome-lysosome fusion. J Cell Sci 130(7):1209–1216
Kuchitsu Y, Fukuda M (2018) Revisiting Rab7 functions in mammalian autophagy: Rab7 Knockout Studies. Cells 7(11)
Jager S et al (2004) Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci 117(Pt 20):4837–4848
Ding X et al (2019) RAB2 regulates the formation of autophagosome and autolysosome in mammalian cells. Autophagy 15(10):1774–1786
Lorincz P et al (2017) Rab2 promotes autophagic and endocytic lysosomal degradation. J Cell Biol 216(7):1937–1947
Lund VK, Madsen KL, Kjaerulff O (2018) Drosophila Rab2 controls endosome-lysosome fusion and LAMP delivery to late endosomes. Autophagy 14(9):1520–1542
Fujita N et al (2017) Genetic screen in Drosophila muscle identifies autophagy-mediated T-tubule remodeling and a Rab2 role in autophagy. Elife 6
Altmann C et al (2016) Progranulin overexpression in sensory neurons attenuates neuropathic pain in mice: Role of autophagy. Neurobiol Dis 96:294–311
Feng T et al (2016) Growth factor progranulin promotes tumorigenesis of cervical cancer via PI3K/Akt/mTOR signaling pathway. Oncotarget 7(36):58381–58395
Saxton RA, Sabatini DM (2017) mTOR signaling in growth, metabolism, and disease. Cell 168(6):960–976
Codogno P, Meijer AJ (2005) Autophagy and signaling: their role in cell survival and cell death. Cell Death Differ 12(Suppl 2):1509–1518
Noda T (2017) Regulation of autophagy through TORC1 and mTORC1. Biomolecules 7(3)
Lorincz, P, Juhasz G (2019) Autophagosome-lysosome fusion. J Mol Biol
Eskelinen EL (2008) To be or not to be? Examples of incorrect identification of autophagic compartments in conventional transmission electron microscopy of mammalian cells. Autophagy 4(2):257–260
Reddy PH (2011) Mitochondrial dysfunction and oxidative stress in asthma: implications for mitochondria-targeted antioxidant therapeutics. Pharmaceuticals (Basel) 4(3):429–456
Sachdeva K et al (2019) Environmental exposures and asthma development: autophagy, mitophagy, and cellular senescence. Front Immunol 10:2787
Garza-Lombo C et al (2020) Redox homeostasis, oxidative stress and mitophagy. Mitochondrion 51:105–117
Kanki T et al (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell 17(1):98–109
Kinya O et al (2015) BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32. Autophgy p. 1932–1933
Gegg ME, Schapira AH (2016) Mitochondrial dysfunction associated with glucocerebrosidase deficiency. Neurobiol Dis 90:43–50
Osellame LD, Duchen MR (2013) Defective quality control mechanisms and accumulation of damaged mitochondria link Gaucher and Parkinson diseases. Autophagy 9(10):1633–1635
Acknowledgements
We thank Drs. Yan Deng and Fengxia Liang at NYU Medical School OCS Microscopy Core for their assistance with confocal and electronic microscope images. We also thank all lab members for the insightful discussions. Patents have been filed by NYU that claim PGRN and its derivatives for the diagnosis and treatment of Gaucher disease (PCT/US2015014364).
Funding
This work was supported partly by NIH research grants R01NS103931, R01AR062207, R01AR061484, and R01AR076900.
Author information
Authors and Affiliations
Contributions
X. Zhao, J. Jian, R. Liberti, and W. Fu designed and performed experiments, collected and analyzed data, and wrote the paper. A. Hettinghouse assisted with experiments and editing the manuscript. Y. Sun contributed to the conceptualization and data interpretation. C. J. Liu supervised this study, analyzed data, and co-wrote and edited the manuscript.
Corresponding author
Ethics declarations
Ethics approval
The full name of the Ethics Committee, from which written approval was obtained allowing to carry out the experiments, is stated in the “Materials and methods” section.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhao, X., Liberti, R., Jian, J. et al. Progranulin associates with Rab2 and is involved in autophagosome-lysosome fusion in Gaucher disease. J Mol Med 99, 1639–1654 (2021). https://doi.org/10.1007/s00109-021-02127-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-021-02127-6