Abstract
Numerous allelopathic plant secondary metabolites impact plant–microorganism interactions by injuring plant-associated beneficial bacteria and fungi. Fungi belonging to the genus Trichoderma positively influence crops, including benzoxazinone-containing maize. However, benzoxazinones and their downstream metabolites such as benzoxazolinone and phenoxazinones are often fungitoxic. Specimen Trichoderma viride F-00612 was found to be insensitive to 100-µM phenoxazinone and 500-µM benzoxazolinone. Screening of 46 additional specimens of ascomycetes revealed insensitivity to phenoxazinones among fungi that cause disease in benzoxazinone-producing cereal crops, whereas many other ascomycetes were highly sensitive. In contrast, most of the screened fungi were insensitive to benzoxazolinone. T. viride F-00612 was associated with bacteria and, thus, existed as a consortium. By contrast, Enterobacter species and Acinetobacter calcoaceticus were prominent in the original specimen, and Bacillus species predominated after antibiotic application. Prolonged cultivation of T. viride F-00612 in liquid medium and on Czapek agar in the presence of < 100 µM phenoxazinone and < 500 µM benzoxazolinone resulted in a massive loss of bacteria accompanied by impacted fungal growth in the presence of phenoxazinone. The original consortium was actively involved in implementing metabolic sequences for the degradation and detoxification of nitrated benzoxazolinone derivatives. The 2-aminophenol was rapidly converted into acetamidophenol, but benzoxazolinone, methoxylated benzoxazolinone, and picolinic acid remained unchanged. Excluding phenoxazinone, none of the tested compounds markedly impaired fungal growth in liquid culture. In conclusion, members of the T. viride F-00612 consortium may contribute to the ability to manage benzoxazinone downstream products and facilitate BOA-6-OH degradation via nitration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Roots exude primary and secondary metabolites, which are essential for establishment of the rhizosphere microbiota (Zwetsloot et al. 2018). The secondary metabolites are particularly important for inter- and intra-species interactions. Recently, several studies have demonstrated the effects of released plant secondary metabolites on bacterial and fungal community structures in agricultural soils and the rhizosphere, demonstrating a modification of the original species diversity and abundances of species. For instance, p-coumaric acid shapes the cucumber rhizosphere microbial community, whereby the Fusarium oxysporum population is increased. Syringic acid has a positive effect on fungal species diversity but reduces the diversity of bacterial species (Zhou and Wu 2012; Wang et al. 2018). The coumarin scopoletin modulates the Arabidopsis root microbiome, thereby favoring plant growth-promoting bacteria while suppressing F. oxysporum (Stringlis et al. 2018). Glucosinolates and their breakdown products dramatically remodel the bacterial community structure (Hu et al. 2014; Siebers et al. 2018) and decrease fungal diversity in monocultures of rapeseed and in culture systems of a canola–wheat rotation (Hilton et al. 2013; Hansen et al. 2019). Salicylic acid modulates the Arabidopsis root microbiome (Lebeis et al. 2015), and benzoxazinoids have been described as modifiers of the soil microbiota (Hu et al. 2018; Kudjordjie et al. 2019). The mentioned compounds are known to function as allelochemicals that affect plant viability. Some of their effects seem to be indirect via influencing plant–microorganism interactions. For instance, allelochemicals can injure associated beneficial bacteria and fungi, leading to damage or even death of the partner plants. The involvement of microorganisms in allelopathic interactions was recently reviewed by Cipollini et al. (2012). Elimination of the original and converted secondary metabolites with allelopathic potential is, therefore, of essential importance to maintain a soil quality that is suitable for crop rotation demanding a high microbial species diversity.
Benzoxazinones are a group of natural bioactive compounds that are synthesized in several Poaceae and some dicotyledonous species (Sicker and Schulz 2002), and several degradation compounds are known, such as the benzoxazolinones BOA and MBOA, and 2-aminophenol and its derivatives. In a recent study, we have shown that the degradation of benzoxazinoids and their downstream metabolites by microorganisms requires a high energy input. Associations between fungal and bacterial species are necessary to orchestrate degradation (Schütz et al. 2019), which likely sometimes occurs in cooperation with plants. Spontaneous oxidative dimerization of 2-aminophenol to biocidal 2-amino-3H-phenoxazin-3-one (APO) is one possible product (Schulz et al. 2013), whereas others depend on the presence of distinct fungal and bacterial species and environmental conditions. Burkholderia xenovorans LB400 degrades 2-aminophenol into fragments that can enter the tricarboxylic acid (TCA) cycle, with picolinic acid as a spontaneous byproduct (Chirino et al. 2013). Heterocyclic cleavage of benzoxazolinone (BOA) is performed by Acinetobacter calcoaceticus, and the resultant 2-aminophenol dimerizes to APO (Massalha et al. 2017). This bacterium is common in soil and is a member of the plant microbiota, for instance, in the rhizosphere of Avena sativa (Andreazza et al. 2012).
Few fungi are known to cleave the BOA heterocycle, a prerequisite action for certain detoxification reactions by fungi and bacteria. For instance, N-(2-hydroxyphenyl) malonamic acid (oHPMA) and 2-acetamidophenol (AAP) are produced from BOA by Fusarium species (Glenn et al. 2001, 2002, 2003; Kettle et al. 2015). Several strains of Gaeumannomyces graminis, another plant pathogen, detoxify BOA and yield the same detoxification products (Friebe et al. 1998). The first step in the pathway is a hydrolysis reaction that creates 2-aminophenol, which is catalyzed by enzymes possessing domain homology to the metallo-β-lactamase superfamily (Glenn et al. 2016). Benzoxazolinones, and particularly the downstream product APO, can be isolated from soil several weeks after wheat culture, suggesting slow degradation (Macias et al. 2005; Fomsgaard et al. 2004). For their own advantage, plants and their associated microorganisms attempt to decontaminate their narrow surroundings from BOA. When exposed to BOA, many plants synthesize BOA-6-O-glucoside as a detoxification product (Sicker et al. 2004). The aglycon BOA-6-OH, which is found at the root surface, presents a substrate for microbial transformation and can be nitrated at position 5. This substitution allows for subsequent microbial degradation. Several soil bacteria isolates, two fungal species (Papulaspora sepedonioides and Trichoderma viride), and an Actinomucor elegans microbial consortium isolated from the Abutilon theophrasti root surface quickly degrade the nitro compound (Schulz et al. 2017a, b, 2018; Schütz et al. 2019).
For decades, Trichoderma species have been used as biological control agents, especially of Fusarium species (Popiel et al. 2008). Trichoderma species are known to possess excellent detoxification capacities, including pathways to detoxify Fusarium toxins (Tian et al. 2017). Trichoderma spp. establish beneficial interactions with crops such as, among others, benzoxazinone-containing maize (Saravanakumar et al. 2017). Because of the agricultural importance of Trichoderma species, we were interested in elucidating the growth behavior of another T. viride specimen during exposure to such compounds.
Here, we investigated the sensitivity towards BOA and APO of T. viride F-00612, a voucher specimen from the Culture Collection of Microscopic Fungi of the Department of Physiology and Taxonomy of Micromycetes of D.K. Zabolotny, Institute of Microbiology and Virology of the National Academy of Sciences of Ukraine, and its ability to degrade downstream metabolites. To gain insights into the sensitivity (or lack thereof) of ascomycetes to the compounds, we screened additional soil fungi from the mentioned collection. We found that root surfaces could be sites of fungal exposure to APO. T. viride F-00612 was also examined for potentially associated bacteria.
Materials and methods
In vitro synthesis of APO and APO degradation
APO was synthesized by catalytic cyclocondensation with horseradish peroxidase (HRP) to yield high-purity preparations (Giurg et al. 2007) (Fig. S1). The preparative assay contained 5 mL of 10-mM 2-aminophenol and 2.5 mL of 17.5-mM H2O2 in 20 mL of 100-mM phosphate buffer, pH 6.8 containing 1 U HRP (Sigma-Merck, Germany). APO production was started within seconds and stopped after 10 min by extraction with EtOAc. The APO-containing organic phase was evaporated, and the residue was dissolved in 100% methanol. A highly concentrated solution was stored at 4 °C for crystallization of the compound. The crystals were washed with water, dried, and used for the experiments.
Oxidative degradation of APO was performed at room temperature in the daylight in macro-cuvettes containing 25-µM APO (final) in 1.9 mL of 50% methanol and 100 µL of H2O2 (1.7; 17 and 170 mM final) or 100 µL of H2O (control). Prior to analysis, 0.5 mL of methanol was added, and the mixtures were vigorously shaken. Aliquots were collected immediately for high-performance liquid chromatography (HPLC) analysis (Fig. S2). Measurements were performed at the start of the reactions and after 3 days.
Growth tests
All fungi were from the Culture Collection of Microscopic Fungi of the Department of Physiology and Taxonomy of Micromycetes of D.K. Zabolotny Institute of Microbiology and Virology of the National Academy of Sciences of Ukraine. A total of 47 strains from 44 species were assessed for tolerance to APO and BOA. APO (100 µM) or BOA (740 µM), both dissolved in methanol, were added to Czapek agar prior to sterilization. Next, 20 mL of sterile medium was poured into Petri dishes (8.5 cm). Controls were mixed with the same amount of organic solvent (0.1%). The growing fungal mycelium was placed at the center of each plate and incubated in the dark at 25 °C. Fungal growth was measured on days 3, 5, 7, and 10. The relative colony diameter after 10 days was compared to the growth of the control, which was defined as 100%. Each experiment was repeated 3 times. Species and strains are listed in Table S1.
In another series of experiments, T. viride was harvested from liquid medium (see below) and cultured on Czapek agar plates with different APO concentrations (100, 210, and 430 µM).
Growth of Trichoderma viride F-00612 in liquid culture supplemented with BOA and derivatives
To grow T. viride, 1 mL of overnight preculture (in YEP medium) was transferred to flasks containing 15 mL of Czapek medium supplemented with 0.5-mM BOA or one of the following compounds: 0.5-mM 2-aminophenol (AP), 0.5-mM MBOA, 0.5-mM BOA-6-OH, 0.5-mM picolinic acid, 100-µM APO, 50-µM NMBOA (6-methoxy-4-nitro-benzoxazolin-2(3H)-one), and 33-µM NBOA-6-OH (6-hydroxy-5-nitrobenzo[d]oxazol-2(3H)-one). The nitrated compounds were also added to liquid cultures containing fungal material from the previous liquid cultures. AP, MBOA, BOA-6-OH, and picolinic acid were purchased from Sigma-Merck, Germany. Nitrated BOA-6-OH and MBOA were synthesized (Schulz et al. 2017a, b, 2018). Growth was monitored every 24 h for a total of 72 h by measuring the OD600 of culture aliquots. HPLC was used to analyze compound conversion and degradation in culture aliquots (Schulz et al. 2017a, b). oHPMA and AAP were gifts from Dieter Sicker, University of Leipzig, and 2-acetylamino-3H-phenoxazin-3-one (AAPO) was a gift from C. Werner, University of Zürich (Zikmundova et al. 2002).
Identification of microorganisms associated with Trichoderma viride F-00612
Trichoderma viride F-00612 was cultivated on malt extract agar (1% or 2%) at 22 °C. The sample biomass was collected after 2 and 9 days of growth. DNA was extracted using CTAB/chloroform extraction. To identify associated microorganisms, touch-down PCR (TD-PCR) was performed using primers for bacteria (27f: AGAGTTTGATCMTGGCTCAG, 519r: GWATTACCGCGGCKGCTG, 533f: GTGCCAGCAGCCGCGGTAA, 1100r: AGGGTTGCGCTCGTTG, 16S.1100.F16: CAACGAGCGCAACCCT, 1492r(s): ACCTTGTTACGACTT), (Lane 1991; Turner et al. 1999) and fungi (ITS1f: CTTGGTCATTTAGAGGAAGTAA, ITS4: TCCTCCGCTTATTGATATGC (Op De Beeck et al. 2014). Fungal primers were used to check the success of the DNA extraction. PCR products were purified using the NucleoSpinGel and PCR Clean-up Kit (Macherey & Nagel, Düren, Germany) and sequenced by GATC Biotech. AG (Konstanz, Germany). For sequencing, 5-µL template DNA (20–80 ng/µL) was mixed with 5 µL of primer (5 pmol/µL). Sequences were searched against the nucleotide database at NCBI using BLASTN. The accession numbers of the 16S rRNA from identified bacteria in untreated T. viride F-00612 included A. calcoaceticus MH915582, Enterobacter cloacae MH915583, and Enterobacter ludwigii MH915584. Identified bacteria that were associated with T. viride F-00612 cultured in the presence of antibiotics included Bacillus subtilis MH915585, B. safensis MH915586, and B. pumilus MH915587 (https://www.ncbi.nlm.nih.gov/nucleotide/).
Variations of Trichoderma viride culture
Trichoderma viride was cultured under the following conditions to identify additional bacterial species and to study the stability of the species composition of the consortium: (1) Czapek agarose (original condition); (2) hyphae from Czapek agarose cultured in liquid Czapek medium; (3) T. viride from liquid culture cultured on Czapek agarose; (4) growing hyphae from Czapek agarose plates cultured on Czapek agarose plates supplemented with 500 µM BOA or 100, 200, or 430 µM APO; or (5) T. viride cultured on 1% malt extract agarose plates supplemented with 100 µg/mL ampicillin, 50 µg/mL kanamycin, 10 µg/mL tetracycline, and 40 µg/mL ciprofloxacin to eliminate associated bacteria. Cultivation in the presence of antibiotics was performed as described by Arendt et al. (2016).
Mycelia scraped from plates or droplets of liquid cultures were used for DNA extraction and TD-PCR as described above. PCR with the fungal primers ITS1f + ITS4 was used to check the isolated DNA.
Statistics
Each data point is based on at least three biological replicates from three independent experiments unless otherwise noted. Error bars are the standard deviation (SD). For statistical analysis of all the data, the t test was used. Data in the figures are presented as the mean ± SD. Significant differences compared with control samples are indicated (t test; *, p < 0.05; **, p < 0.005; ***, p < 0.0005).
Results
Relevance of fungal exposure to APO
Root exudation of benzoxazinone (BX) by cereals such as rye or maize leads to benzoxazolinone accumulation at the surface of young roots and within the rhizosphere. Benzoxazolinone heterocycle cleavage is performed by A. calcoaceticus and fungi such as Fusarium species, yielding 2-aminophenol as an important intermediate. As shown for HRP, 2-aminophenol can be a substrate for plant peroxidases. Under natural conditions, root surface peroxidases may contribute to oxidative 2-aminophenol dimerization and APO precipitation at the root surface. Consequently, incubation of seedlings with 2-aminophenol or BOA results in yellow–orange-colored roots (Fig. S3) (Sánchez-Moreiras and Reigosa 2005; Schulz and Wieland 1999).
Thus, the root surface of BX-releasing species and their rhizosphere could indeed be sites where root-associated fungi contact APO. Fungal APO insensitivity or sensitivity is relevant when the conversion of 2-aminophenol into APO occurs more rapidly than its detoxification or degradation.
Benzoxazolinones, downstream metabolites, and APO
Trichoderma viride F-00612 was not affected by APO up to concentrations of 100 µmol when grown on plates (Fig. 5). We tested APO and the benzoxazolinones MBOA and BOA, the plant detoxification intermediate BOA-6-OH, the nitroaromatic compounds NBOA-6-OH and NMBOA (Schulz et al. 2017b, 2018), 2-aminophenol, and picolinic acid to evaluate their effects on the growth of T. viride F-00612 when cultivated in liquid medium. In liquid culture, T. viride increasingly clumped only in the presence of APO (Fig. 1).
Because of clumping, growth monitoring of the APO sample was stopped after 48 h. Notably, the APO medium contained only traces of AAPO after 72 h (Fig. 2, blue frame). Some fungi transform APO to AAPO and 2-(N-hydroxy) acetylamino-3H-phenoxazin-3-one, which are both more hydrophilic than APO and probably less toxic (Zikmundova et al. 2002).
None of the other compounds had a significant inhibitory effect. HPLC analyses indicated that picolinic acid and BOA (0.5 mM) were not converted. We detected traces of NMBOA in the MBOA sample (Fig. 2, purple frame). The traces were not degraded. In contrast, when the culture medium was supplemented with 50-µM NMBOA, the compound was degraded without identifiable intermediates. BOA-6-OH was nitrated within 48 h to NBOA-6-OH, presenting a degradation intermediate that was further degraded. Small amounts of unidentified compounds were detected (Fig. 2, pink frame).
However, when T. viride was incubated with BOA-6-OH after prolonged liquid culture, almost no NBOA-6-OH was produced, and no degradation of NBOA-6-OH and NMBOA occurred. The 2-aminophenol was mainly detoxified by acetylation, yielding acetamidophenol (AAP), which was not degraded (Fig. 2, green frame).
Since the presence of APO in liquid culture resulted in clumping of the hyphae, fungal material from those cultures was grown on Czapek agar plates with different APO concentrations (100, 210, and 430 µM). We expected a different growth behavior than previously observed, and indeed, we found that no spores developed in the presence of APO, even at the lowest concentration, and increasingly clumpy hyphae indicated impaired growth (Fig. 3). The altered behavior of the fungus when cultured previously in liquid medium prompted us to evaluate the specimen for associated microorganisms.
Picture of Czapek agar plate sections with Trichoderma viride F-00612 grown after previous liquid culture. a No APO resulted in dense hyphae with spores (arrow). b With 100-µM APO, hyphae are less dense and do not have spores. c With 210-µM APO, hyphae start clump (arrow) in some areas and transparent zones are present (arrow); d With 430-µM APO, hyphal growth is strongly inhibited and clumps form (arrows)
Bacteria associated with Trichoderma viride F00612
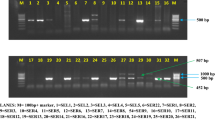
Sequence analysis of the PCR amplification products obtained with bacteria-specific primers revealed that the original T. viride specimen F-00612 was associated with several bacteria (Fig. 4, panel 2A, B, C). Gram-negative E. ludwigii, E. cloacae, and A. calcoaceticus were the most abundant species under the applied culture conditions on Czapek agar plates.
Row (1) cultivation of T. viride F-00612 in the presence of BOA and APO reduces or eliminates associated bacteria. DNA samples from T. viride (cultured on Czapek agar plates) were used. 1A: Samples were cultured with 500-µM BOA (lanes 2–3), 100-µM APO (lanes 4–5), or 200-µM APO (lanes 6–7). In 1B, samples were cultured with 430-µM APO (lanes 8–9). In these samples, all primers were for bacterial DNA. Primers included the following: A2–B9, l 27f and 519r primers; B10–11, 533f and 1100r primers; B12–13, 16S.1100.F16 and 1492r(s) primers. In 1C and 1D, fungal ITS1f and ITS4 primers were used. Samples were cultured with 500-µM BOA (lanes 14–17, 100-µM APO (lanes 18–19); 200-µM APO (lanes 20–21); negative control (lanes 22–23), or 430-µM APO (lane 24). Row 2) Bacteria detected in T. viride F00612. PCR with was performed with DNA samples of T. viride F-00612 grown on 1% malt extract agar media (lanes 2–5). 2A utilizes bacterial primers 16S.1100.F16 and 1492r(s), 2B utilizes bacterial primers 533f and 1100r, and 2C uses bacterial primers 27f and 519r. Row (3) Gel 3: culture in liquid medium severely reduces or eliminates associated bacteria. TD-PCR with DNA from T. viride F-00612 cultured in liquid Czapek medium. Lanes 2, 7, and 10 are empty. Primers included ITS1f and ITS4 (lanes 3–4), 27f and 519r (lanes 5–6), 533f and 1100r (lanes 8–9), and 16S.1100.F16 and 1472r(s) (lanes 11–12). Row (3), Gel 4: products obtained with bacterial primers indicated the presence of bacteria despite antibiotics. TD-PCR with DNA from T. viride cultured on 2% malt extract agar with antibiotics. Primers included ITS1f and ITS4 (lanes 2–3), 27f and 519r (lanes 4–5), 533f and 1100r (lanes 6–7), 16S.1100.F16 and 1483r(s) (lanes 8–9), ITS1f and ITS4 (lanes 10–11), and 27f and 519r (lanes 12–13). In all panels, lane 1 contains a 1-kD ladder
An approach to eliminate the bacterial consortium of T. viride with antibiotics failed, as other bacteria could be identified after culturing in the presence of the antibiotics (Fig. 4, panel 4). However, we were unable to isolate DNA from the previously identified bacterial species. Following antibiotic treatment, gram-positive Bacillus species, which were not detected previously in the mycelium, were most abundant (B. pumilis, B. subtilis, and B. safensis). The biodiversity of the microbial consortium was massively reduced during continued cultivation in the liquid medium. Small amounts of bacterial DNA were obtained, and PCR products were insufficient for sequencing analyses (Fig. 4, panel 3). Reactivation of bacterial growth by transferring the mycelium from liquid cultures to Czapek agar plates was not possible. A loss of bacteria over time was also observed on plates supplemented with APO or BOA (Fig. 4, panel 1 A–C). Thus, the allelochemicals and liquid culturing ultimately led to a continual loss of the associated bacteria, which affected the degradation of the nitro compounds and altered the sensitivity towards APO, in comparison to growth of the original consortium on Czapek agar plates in the presence of 100-µM APO (see Fig. 5).
Growth of T. viride and further fungi in the presence of APO and BOA
To evaluate the growth behavior of T. viride in the presence of APO and BOA in comparison to other fungi, we tested the growth of 47 other voucher specimens, including numerous Fusarium and Penicillium species, on Czapek agar plates. Most of the tested Fusarium and Penicillium species were not sensitive or were barely sensitive to APO (Fig. 5). The Alternaria state of Pleospora infectoria (F-01088) exhibited better growth. F. culmorum (F-00121), a species found on BX-containing cereals, was moderately inhibited by ~ 30%. Insensitive, less sensitive, and sensitive Aspergillus species were found. All tested Mucor species were sensitive. No pathogen of BX-containing cereals belonging to the Fusarium genus was considerably sensitive to APO with the exceptions of F. graminearum and F. culmorum. Among the group of pathogenic fungi with lower plant host specificity, only one species (F. solani F-00530) was not sensitive. The tested plant endophytes were sensitive (Nigrospora oryzae F-00202 and Talaromyces stipitatus F-00203). The group of plant growth-promoting fungi and those used for biocontrol were composed of insensitive, sensitive, and highly sensitive species (for instance, Cladosporium cladosporioides). The two Trichoderma species and F. incarnatum were insensitive. The two specimens of Penicillium brevicompactum (2.1, 2.2) and Curvularia lunata (22.1; 22.2) differed markedly in their sensitivity to APO. Further information on the tested species is provided in Table S1.
Almost all the tested species were insensitive to BOA or showed low growth inhibition (Fig. 5). Only six species were severely affected, showing 40–50% or more growth inhibition (Cladosporon herbarum, Phoma eupyrena, Mucor hiemalis, Umbelopsis isabellina, Chaetomium globosum, and Paecilomyces variotii). Excluding Chladosprium herbarum, which was more sensitive to BOA than to APO, all the BOA-sensitive fungi were also among the most sensitive ones to APO. Similar to T. viride, some of the tested specimens might harbor bacteria; however, such analyses are beyond the scope of the present study.
Discussion
Coping with BOA downstream metabolites
The present study provides evidence that the consortium T. viride F-00612 nitrates BOA-6-OH and degrades the nitroaromatic compound. In a recent study, another strain of T. viride (BioProject database ID: PRJNA376094, Siebers et al. 2018) was able to degrade NBOA-6-OH but was unable to nitrate BOA-6-OH (Schütz et al. 2019). The Abutilon theophrasti root-associated Actinomucor elegans consortium, which includes the easily culturable bacterial species Pantoea ananatis, Stenotrophomonas maltophilia, and the yeast Papiliotrema baii, nitrates BOA-6-OH and degrades the nitro compound. Culture of isolated P. ananatis in liquid Czapek medium containing BOA-6-OH has revealed that this species contributes to the degradation of this compound via nitration (Schulz et al. 2017b). Thus, nitration of BOA-6-OH is widespread and seems to be a prerequisite for subsequent degradation by microorganisms. Because of the methoxy group, nitration of MBOA occurs at low rates, and direct degradation of MBOA by all the presently tested microorganisms does not seem likely (Schütz et al. 2019; Schulz et al. 2018). Nitroaromatic compound degradation by certain bacteria was investigated with synthetic compounds originating from anthropogenic activities that contaminate the environment. Depending on the conditions and bacterial species, oxidative and reductive pathways can be performed, yielding either 2-hydroxymuconic semialdehyde or 2-aminomuconic semialdehyde and their derivatives, respectively (Ju and Parales 2010).
The tested T. viride specimens do not degrade BOA but rather convert AP into AAP (Schütz et al. 2019, this study). Acetylation of AP avoids the generation of growth-reducing APO. Growth reduction by APO may be less severe when the compound is converted into AAPO, which was not tested in this study because the conversion to AAPO seemed to be very slow under the applied liquid culture conditions (Fig. 4).
Our results indicate that fungal and bacterial associations broaden the detoxification and degradation capacities for BOA-6-OH via nitration and accelerate the conversion of AP into AAP. The assembly of microbial consortia, therefore, increases the probability of survival and perhaps uncovers additional colonization sites.
Sensitivity of ascomycetes to APO and BOA
When cultivated on Czapek agar, the APO sensitivity of all ascomycetes screened in this study mirrored the ecological niche and lifestyle of the studied fungi. All Fusarium species infesting predominantly BX-producing cereals could be grouped as highly insensitive species, whereas other Fusarium species with lower host specificity were more sensitive. A possible explanation for the insensitivity to lower APO concentrations could be the destruction of APO by oxidative processes. Fungi produce high concentrations of extracellular H2O2 and other reactive oxygen species via NADPH oxidase (Hansel et al. 2012). The addition of H2O2 to APO solutions was sufficient to destroy the compound without enzymatic support (Fig. S2, Fig. S3). We assume that many fungi can destroy APO oxidatively through the extracellular release of superoxide and increased H2O2 concentration in the narrow surroundings of hyphae. Efficient protection of the mycelium against such harsh conditions may contribute to APO insensitivity. Another strategy to escape APO toxicity is the degradation or polymerization of the precursor compound 2-aminophenol via laccase-like multicopper oxidase (LMCO), although we found no evidence for polymerization in the HPLC runs. However, Levasseur et al. (2010) have demonstrated that LMCO genes are present in many ascomycetes, including Trichoderma species. A defined role for extracellular enzymes, such as laccases and peroxidases, has still to be elucidated. Avoidance of AP generation, a key step for subsequent APO production, may be another strategy. Growth of Fusarium species with BOA in our study aligns with the published results reported by Glenn et al. (2016) and Kettle et al. (2015) and can be explained by the previously described detoxification pathways for BOA. Because many other ascomycetes are also insensitive to the allelochemical, bacterial associations should be considered for these specimens. Moreover, insensitive species may have transporters for the efflux of absorbed BOA, similar to the ABC transporter of Botrytis cinerea that enhances tolerance to camalexin (Stefanato et al. 2009).
Identified members of the consortium: properties and fate
The species composition of bacteria associated with T. viride F-00612 depends on the culture conditions. Culture of T. viride in liquid medium supplemented with antibiotics or the allelochemicals BOA and APO led to an altered species composition of the consortium and, depending on the concentration, a dramatic reduction in species abundance, perhaps due to loss of the associated bacteria. Because the loss of those bacteria increases the sensitivity of the fungus to APO, the bacteria seemed to improve the tolerance of T. viride to APO when present at low concentrations by as yet unknown mechanisms. Although BOA-degrading A. calcoaceticus was identified in association with T. viride, no APO generation occurred on agar plates supplemented with BOA, the reason for which is unclear. We speculate that the fungus may suppress BOA degradation via changes in the physiological state of the associated A. calcoaceticus. The distribution of the bacteria within the mycelium may also restrict degradation reactions to areas of richer bacterial colonization.
The mycosphere is defined as a habitat harboring different bacteria that are involved in highly complex interactions. Bacterial density is much higher in the mycosphere than in the surrounding soil. The mycosphere is also believed to present a space that is especially suitable for accelerated horizontal gene transfer, driving microbial evolution (Zhang et al. 2014; Berthold et al. 2016). Some bacteria associated with T. viride F-00612 may be integral components of the mycosphere that was collected with the fungus, while others may be endophytes. Endophytic and rhizosphere-colonizing strains of E. ludwigii, a species of the E. cloacae complex, have plant growth-promoting properties (Yousaf et al. 2011). It is known that associated bacteria dramatically influence the lifestyle of fungi, even transforming a plant pathogen to a beneficial, plant growth-promoting fungus (Minerdi et al. 2011; Bitas et al. 2015). The T. viride F-00612 specimen is a natural consortium that harbors numerous bacteria with plant growth-promoting and other valuable properties, such as amelioration of the impact of allelochemicals. These properties may help to potentiate the plant-protecting effects of T. viride.
References
Andreazza R, Okeke BC, Pieniz S, Camargo FA (2012) Characterization of copper-resistant rhizosphere bacteria from Avena sativa and Plantago lanceolata for copper bioreduction and biosorption. Biol Trace Elem Res 146:107–115. https://doi.org/10.1007/s12011-011-9228-1
Arendt KR, Hockett KL, Araldi-Brondolo SJ, Baltrus DA, Arnold AE (2016) Isolation of endohyphal bacteria from foliar ascomycota and in vitro establishment of their symbiotic associations. Appl Environ Microbiol 82:2943–2949. https://doi.org/10.1128/aem.00452-16
Berthold T, Centler F, Hübschmann T, Remer R, Thullner M, Harms H, Wick LY (2016) Mycelia as a focal point for horizontal gene transfer among soil bacteria. Sci Rep 6:36390. https://doi.org/10.1038/srep36390
Bitas V, McCartney N, Li N, Demers J, Kim J-E, Kim H-S, Brown KM, Kang S (2015) Fusarium oxysporum volatiles enhance plant growth via affecting auxin transport and signaling. Front Microbiol 6:1248. https://doi.org/10.3389/fmicb.2015.01248
Chirino B, Strahsburger E, Agulló L, González M, Seeger M (2013) Genomic and functional analyses of the 2-aminophenol catabolic pathway and partial conversion of its substrate into picolinic acid in Burkholderia xenovorans LB400. PLoS One 8(10):e75746. https://doi.org/10.1371/journal.pone.0075746
Cipollini D, Rigsby CM, Barto EK (2012) Microbes as targets and mediators of allelopathy in plants. J Chem Ecol 38:714–727. https://doi.org/10.1007/s10886-012-0133-7
Fomsgaard IS, Mortensen AG, Carlsen SCK (2004) Microbial transformation products of benzoxazolinone and benzoxazinone allelochemicals—a review. Chemosphere 54:1025–1038
Friebe A, Vilich V, Hennig L, Kluge M, Sicker D (1998) Detoxification of benzoxazolinone allelochemicals from wheat by Gaeumannomyces graminis var. tritici, G. graminis var. graminis, G. graminis var. avenae, and Fusarium culmorum. Appl Environ Microbiol 64:2386–2391
Giurg M, Piekielska K, Gębala M, Ditkowski B, Wolański M, Peczyńska-Czoch W, Młochowski J (2007) Catalytic oxidative cyclocondensation of o-aminophenols to 2-amino-3H-phenoxazin-3-ones. Synth Commun 37:1779–1789. https://doi.org/10.1080/00397910701316136
Glenn AE, Hinton DM, Yates IE, Bacon CW (2001) Detoxification of corn antimicrobial compounds as the basis for isolating Fusarium verticillioides and some other Fusarium species from corn. Appl Environ Microbiol 67:2973–2981
Glenn AE, Gold SE, Bacon CW (2002) Fdb1 and Fdb2, Fusarium verticillioides loci necessary for detoxification of preformed antimicrobials from corn. Mol Plant Microbe Interact 15:91–101
Glenn AS, Meredith FI, Morrison WH III, Bacon CW (2003) Identification of intermediate and branch metabolites resulting from biotransformation of 2-benzoxazolinone by Fusarium verticillioides. Appl Environ Microbiol 69:3165–3169
Glenn AE, Davis CB, Gao M, Gold SE, Mitchell TR, Proctor RH, Stewart JE, Snook ME (2016) Two horizontally transferred xenobiotic resistance gene clusters associated with detoxification of benzoxazolinones by Fusarium species. PLoS One 11:e0147486. https://doi.org/10.1371/journal.pone.0147486
Hansel CM, Zeiner CA, Santelli CM, Webb SM (2012) Mn(II) oxidation by an ascomycete fungus is linked to superoxide production during asexual reproduction. PNAS 109:12621–12625
Hansen JC, Schillinger WF, Sullivan TS, Paulitz TC (2019) Soil microbial biomass and fungi reduced with canola introduced into long-term monoculture wheat rotations. Front Microbiol 10:1488
Hilton S, Bennett AJ, Keane G, Bending GD, Chandler D, Stobart R, Mills R (2013) Impact of shortened crop rotation of oilseed rape on soil and rhizosphere microbial diversity in relation to yield decline. PLoS One 8(4):e59859. https://doi.org/10.1371/journal.pone.0059859PubMed PMID: 23573215
Hu P, Hollister EB, Somenahally AC, Hons FM, Gentry TJ (2014) Soil bacterial and fungal communities respond differently to various isothiocyanates added for biofumigation. Front Microbiol 5:729
Hu L, Robert CAM, Cadot S, Zhang X, Ye M, Li B et al (2018) Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat Commun 9:2738. https://doi.org/10.1038/s41467-018-05122-7
Ju KS, Parales RE (2010) Nitroaromatic compounds, from synthesis to biodegradation. Microbiol Mol Biol Rev 74:250–272. https://doi.org/10.1128/MMBR.00006-10
Kettle AJ, Batley J, Benfield AH, Manners JM, Kazan K, Gardinera DM (2015) Degradation of the benzoxazolinone class of phytoalexins is important for virulence of Fusarium pseudograminearum towards wheat. Mol Plant Pathol 16:946–962
Kudjordjie EN, Sapkota R, Steffensen SK, Fomsgaard IS, Nicolaisen M (2019) Maize synthesized benzoxazinoids affect the host associated microbiome. Microbiome 7:59. https://doi.org/10.1186/s40168-019-0677-7
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–175
Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, Mcdonald M, Malfatti S, Del Rio TG, Jones CD, Tringe SG, Dangl JL (2015) Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349(6250):860–864
Levasseur A, Saloheimo M, Navarro D, Andberg M, Pontarotti P, Kruus K, Record E (2010) Exploring laccase-like multicopper oxidase genes from the ascomycete Trichoderma reesei: a functional, phylogenetic and evolutionary study. BMC Biochem 11:32. https://doi.org/10.1186/1471-2091-11-32
Macias FA, Oliveros-Bastidas A, Marin D, Castellano D, Simonet AM, Molinillo JMG (2005) Degradation studies on benzoxazinoids. Soil degradation dynamics of (2R)-2-ß-D-glucopyranosyl-4-hydroxy(2H)-1,4-phenoxazin-3-(4H)-one (DIBOA-Glc) and its degradation products, phytotoxic allelochemicals from Gramineae. J Agric Food Chem 53:538–548
Massalha H, Korenblum E, Tholl D, Aharon A (2017) Small molecules below-ground: the role of specialized metabolites in the rhizosphere. Plant J 90:788–807
Minerdi D, Bossi S, Maffei ME, Gullino ML, Garibaldi A (2011) Fusarium oxysporum and its bacterial consortium promote lettuce growth and expansin A5 gene expression through microbial volatile organic compound (MVOC) emission. FEMS Microbiol Ecol 76:342–351. https://doi.org/10.1111/j.1574-6941.2011.01051.x
Op De Beeck M, Lievens B, Busschaert P, Declerck S, Vangronsveld J, Colpaert JV (2014) Comparison and validation of some ITS primer pairs useful for fungal metabarcoding studies. PLoS One. https://doi.org/10.1371/journal.pone.0097629(Published 2014 Jun 16)
Popiel D, Kwaśna H, Chełkowski J, Stępień L, Laskowska M (2008) Impact of selected antagonistic fungi on Fusarium species—toxigenic cereal pathogens. Acta Mycol 43:29–40
Sánchez-Moreiras AM, Reigosa MJ (2005) Whole plant response of lettuce after root exposure to BOA (2(3H)-benzoxazolinone). J Chem Ecol 31:2689–2703. https://doi.org/10.1007/s10886-005-7620-z
Saravanakumar K, Li Y, Yu C, Wang QQ, Wang M, Sun J et al (2017) Effect of Trichoderma harzianum on maize rhizosphere microbiome and biocontrol of Fusarium stalk rot. Sci Rep 7:1771. https://doi.org/10.1038/s41598-017-01680-w
Schulz M, Wieland I (1999) Variations in metabolism of BOA among species in various field communities—biochemical evidence for co-evolutionary processes in plant communities? Chemoecology 9:133–141
Schulz M, Tabaglio V, Marocco A, Macias FA, Molinillo JMG (2013) Benzoxazinoids in rye allelopathy—from discovery to application in sustainable weed control and organic farming. J Chem Ecol 39:154–174
Schulz M, Sicker D, Schackow O, Hennig O, Yurkov A, Siebers M, Hofmann D, Disko U, Ganimede C, Mondani L, Tabaglio V, Marocco A (2017a) Cross-cooperations of Abutilon theophrasti Medik and root surface colonizing microorganisms disarm phytotoxic hydroxy-benzoxazolin-2(3H)-ones. Plant Signal Behav. https://doi.org/10.1080/15592324.2017.1358843
Schulz M, Sicker D, Schackow O, Hennig L, Hofmann D, Disko U, Ventura M, Basyuk K (2017b) 6-Hydroxy-5-nitrobenzo[d]oxazol-2(3H)-one—a degradable derivative of natural 6-hydroxybenzoxazolin-2(3H)-one produced by Pantoea ananatis. Commun Integr Biol 10:e1302633. https://doi.org/10.1080/19420889.2017.1302633
Schulz M, Hofmann D, Sicker D, Hennig L, Schütz V, Preusche M, Thiele B (2018) Pantoea ananatis converts MBOA to 6-methoxy-4-nitro-benzoxazolin-2(3H)-one (NMBOA) for cooperative degradation with its native root colonizing microbial consortium. Nat Prod Com 13:1271–1275
Schütz V, Bigler L, Girel S, Laschke L, Sicker D, Schulz M (2019) Conversions of benzoxazinoids and downstream metabolites by soil microorganisms. Front Ecol Evol. https://doi.org/10.3389/fevo.2019.00238
Sicker D, Schulz M (2002) Benzoxazinones in plants: occurrence, synthetic access and biological activity. In: Atta-ur-Raman (ed) Studies in natural product chemistry, Bioactive Natural Products Part H, vol 27. Elsevier, Amsterdam, pp 185–232
Sicker D, Hao H, Schulz M (2004) Benzoxazolin-2-(3H)-ones. Generation, effects and detoxification in the competition among plants. In: Macias FA, Galindo JCG, Molinillo JMG, Cutler HG (eds) Recent advances on allelopathy, vol II. CRC Press, Boca Raton
Siebers M, Rohr T, Ventura M, Schütz V, Thies S, Kovacic F, Jaeger KE, Berg M, Dörmann P, Schulz M (2018) Disruption of microbial community composition and identification of plant growth promoting microorganisms after exposure of soil to rapeseed-derived glucosinolates. PLoS One 13(7):e0200160. https://doi.org/10.1371/journal.pone.0200160
Stefanato FL, Abou-Mansour E, Buchala A, Kretschmer M, Mosbach A, Hahn M, Bochet CG, Métraux J, Schoonbeek H (2009) The ABC transporter BcatrB from Botrytis cinerea exports camalexin and is a virulence factor on Arabidopsis thaliana. Plant J 58:499–510. https://doi.org/10.1111/j.1365-313X.2009.03794.x
Stringlis IA, Yu K, Feussner K, Jonge R, Van Bentum S, Van Verk MC, Berendsen RL, Bakker PAHM, Feussner I, Pieterse CMJ (2018) MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. PNAS. https://doi.org/10.1073/pnas.1722335115
Tian Y, Tan Y, Yan Z, Liao Y, Chen J, De Boevre M, De Saeger S, Wu A (2017) Antagonistic and detoxification potentials of Trichoderma isolates for control of zearalenone (ZEN) producing Fusarium graminearum. Front Microbiol 8:2710. https://doi.org/10.3389/fmicb.2017.02710
Turner S, Pryer KM, Miao VP, Palmer JD (1999) Investigating deep phylogenetic relationships among Cyanobacteria and plastids by small subunit rRNA sequence analysis1. J Eukaryot Microbiol 46:327–338. https://doi.org/10.1111/j.1550-7408.1999.tb04612.x
Wang Z, Zhang J, Wu F, Zhou X (2018) Changes in rhizosphere microbial communities in potted cucumber seedlings treated with syringic acid. PLoS One 13(6):e0200007. https://doi.org/10.1371/journal.pone.0200007
Yousaf S, Afzal M, Reichenauer TG, Brady CL, Sessitsch A (2011) Hydrocarbon degradation, plant colonization and gene expression of alkane degradation genes by endophytic Enterobacter ludwigii strains. Environ Pollut 159:2675–2683. https://doi.org/10.1016/j.envpol.2011.05.031Epub 2011 Jun 23
Zhang M, Visser S, Pereira ESMC, van Elsas JD (2014) IncP-1 and PromA group plasmids are major providers of horizontal gene transfer capacities across bacteria in the mycosphere of different soil fungi. Microb Ecol. https://doi.org/10.1007/s00248-014-0482-6
Zhou X, Wu F (2012) p-Coumaric acid influenced cucumber rhizosphere soil microbial communities and the growth of Fusarium oxysporum f. sp. cucumerinum Owen. PLoS One 7(10):e48288. https://doi.org/10.1371/journal.pone.0048288
Zikmundova M, Drandarov K, Hesse M, Werner C (2002) Hydroxylated 2-amino-3H-phenoxazin-3-one derivatives as products of 2-hydroxy-1,4-benzoxazin-3-one (HBOA) biotransformation by Chaetosphaeria sp., an endophytic fungus from Aphelandra tetragona. Zeitschr Naturforsch 57C:660–665
Zwetsloot MJ, Kessler A, Bauerle TL (2018) Phenolic root exudate and tissue compounds vary widely among temperate forest tree species and have contrasting effects on soil microbial respiration. New Phytol 218:530–541. https://doi.org/10.1111/nph.15041
Acknowledgements
Open Access funding provided by Projekt DEAL. M.S., N.V. and A.G. thank the BMBF, Internationales Büro for financial support of part of the study. The authors also thank Dr. Iryna Kurchenko (D.K. Zabolotny Institute of Microbiology and Virology, National Academy of Sciences of Ukraine) for the fungal cultures, Mrs. Esperance Koka Bokaa for help with the T. viride cultures, Scarlett Geunes-Boyer and Cecilia Wall for help with the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Günther Raspotnig.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Voloshchuk, N., Schütz, V., Laschke, L. et al. The Trichoderma viride F-00612 consortium tolerates 2-amino-3H-phenoxazin-3-one and degrades nitrated benzo[d]oxazol-2(3H)-one. Chemoecology 30, 79–88 (2020). https://doi.org/10.1007/s00049-020-00300-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00049-020-00300-w