Abstract
The appY gene has been characterised as conferring resistance to a novel series of antimicrobial benzimidazole derivatives in E. coli MC1061 cells when expressed in high copy-number. A microarray approach was used to identify genes involved in the mechanism of appY-mediated antibacterial resistance, that were up- or down-regulated following induction of the gene in the appY knockout strain JW0553. In total, expression of 90 genes was induced and 48 repressed greater than 2.5-fold (P < 0.05), 45 min after appY induction. Over half the genes up-regulated following appY expression had confirmed or putative roles in acid resistance (AR) and response to oxidative and antibiotic stresses. These included the genes for MdtE and MdtF, which form a multi-drug transporter with TolC and have been implicated in resistance to several antibiotics including erythromycin. Amongst the acid resistance genes were gadAB and adiAC encoding the glutamate-dependant (AR2) and arginine-dependant (AR3) acid resistance systems respectively, in addition to the transcriptional activators of these systems gadE and gadX. In agreement with earlier studies, appA, appCB and hyaA-F were also up-regulated following induction of appY. This study has also confirmed that over-expression of mdtEF confers resistance to these antibacterial benzimidazoles, indicating that the observation of appY conferring resistance to these compounds, proceeds through an appY-mediated up-regulation of this efflux transporter. To assess the importance of the AppY enzyme to acid stress responses, the percentage survival of bacteria in acidified media (pH ≤ 2) was measured. From an initial input of 1 × 106 CFU/ml, the wild-type strain MG1655 showed 7.29% and 0.46% survival after 2 and 4 h, respectively. In contrast, strain JW0553 in which appY is deleted was completely killed by the treatment. Transformation of JW0553 with a plasmid carrying appY returned survival to wild-type levels (7.85% and 1.03% survival at 2 and 4 h). Further dissection of the response by prior induction of each of the three AR systems has revealed that AR1 and AR3 were most affected by the absence of appY. This work highlights an important and previously unidentified role for the AppY enzyme in mediating the responses to several stress conditions. It is likely that the appY gene fits into a complex transcriptional regulatory network involving σS and gadE and gadX. Further work to pinpoint its position in such a hierarchy and to assess the contribution of appY to oxidative stress responses should help determine its full significance. This work is also consistent with recent studies in C. difficile showing that the mechanism of action of ridinilazole involves AT-rich DNA minor groove binding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
A novel series of symmetric head-to-head bis-benzimidazole (BBZ) derivatives was first reported in 1997 [1]. The underlying head-to-head bis-benzimidazole concept was designed by computer modelling as part of a project to develop drug-like DNA sequence reading compounds. The modelling suggested that this BBZ chemotype could span four AT base pairs in duplex DNA, and thus inhibit transcription of AT-rich genes. These predictions were confirmed by DNA foot-printing, surface plasmon resonance binding [2] and X-ray crystallographic studies [1, 3] on two dodecanucleotide duplex complexes (PDB id 1FTD and 453D). Several compounds in the series, with pendant cationic alkylamino groups, also showed cell growth inhibition in a panel of cancer cell lines, as well as modest anti-tumour activity in xenograft models for human cancers [1, 3, 4]. Derivatives with uncharged and more hydrophobic end-groups were devoid of activity. A patent for potential use of the active compounds as anti-cancer agents was granted [5], but ultimately abandoned as it was judged that the lead compound activity was too modest to have beneficial clinical effects in humans.
Several compounds in the series that did not possess cationic charge have subsequently shown anti-bacterial activity, especially against a panel of methicillin resistant Staphylococcus aureus strains and MRSA clinical isolates [6, 7]. There was no significant activity against Gram-negative bacteria. Subsequently in collaboration with Summit Therapeutics PLC, activity was found against the nosocomial pathogen Clostridium difficile (C. difficile) [8]. Their medicinal chemistry programme then discovered a lead compound, the bis-pyridyl derivative, initially termed SMT19969, now named ridinilazole, which has shown remarkable activity and selectivity in cellular and in vivo models and is currently in Phase III clinical evaluation [9,10,11,12].
The mode of action of ridinilazole against C. difficile has not been established until recently. Studies [13] of its interactions with duplex DNA have indicated that AT-rich minor groove DNA is a primary target, in accord with the initial design hypothesis for the compound series and is consistent with the high AT content of the C. difficile genome. We suggest that this is consistent with the results presented here on the mechanism of resistance of ridinilazole and a selected sub-set of bis-benzimidazole derivatives, against several bacterial lines, using standard MIC-type assays together with a genetics study using a multi-copy suppression strategy. This strategy works on the principle that if multiple copies of the molecular target are supplied the antibiotic will be effectively titrated out, enabling growth on otherwise inhibitory concentrations of the antibacterial compound and has been used previously to identify molecular targets of novel antibacterial agents [14]. We report also on the identification of AppY (either at the gene or protein levels) as a key player in the resistance of these compounds in E. coli.
AppY is an AraC family transcriptional regulator in Escherichia coli that is known to control 3 sets of genes with roles in energy metabolism: appA, which encodes acid phosphatase pH 2.5, appCB that produce cytochrome bd-II oxidase and the hyaA-F operon, which encodes hydrogenase-1 [15,16,17]. More recently, appY was identified in a genomic screen for genes that stabilised the stationary phase sigma factor, σS [18], itself a master transcriptional regulator of the metabolic changes associated with entry to stationary phase and the general stress response. It is currently unknown what additional genes might be under control of AppY. In the study describing its original isolation, AppY over-expression was shown to alter the rate of synthesis of at least 30 proteins [15], indicating that additional targets remain to be discovered.
Results
Assessment of BBZ compounds as inhibitors of E. coli growth
We have reported previously on members of this group of symmetric bis-benzimidazoles (BBZ) that have potent inhibitory activity against S. aureus. These compounds were shown to possess inhibitory activity against DNA gyrase, which was presumed to account for their mechanism of action [6]. Here the ability of these compounds to inhibit the growth of E. coli was assessed. No inhibitory activity was observed for any BBZ compound against the K-12 reference strains NCTC 10418 and MG1655 (Table 1). However, the pattern of BBZ activity observed against S. aureus was restored in MC1061. This strain has been reported as being hyper-permeable to small molecules [19], indicating that the lack of activity in most strains is a result of poor penetration across the outer membrane. Time-kill kinetics and scanning electron microscopy revealed that the three most active compounds (BBZ4, BBZ5 and BBZ9) are bacteriostatic and induced pronounced filamentation in MC1061 (Supplementary Fig. S1). These data strongly suggested that the antibacterial activity of these compounds in E. coli would proceed, as in S. aureus, through inhibition of bacterial type-II topoisomerases. However, topoisomerase inhibition assays using purified E. coli enzymes has revealed that of the antibacterial BBZ compounds, only BBZ5 showed even moderate inhibitory activity against DNA gyrase-mediated supercoiling and against topoisomerase IV-mediated decatenation and relaxation activities (Table 1, Supplementary Fig. S2).
Isolation of appY as a multi-copy suppressor of BBZ-mediated growth inhibition
The above results led to a search for other potential targets of the BBZ compounds in E. coli. To this end multi-copy suppression was utilised to identify genes which, when expressed at an artificially high copy number, resulted in resistance to the BBZ compounds. This approach has been used to successfully identify targets of small molecule anti-bacterial compounds [19].
A random genomic library of 3–4 kb fragments of genomic DNA isolated from E. coli strain MC1061 was constructed in the high-copy expression vector pUC19. Following transformation with pUC19 harbouring random genomic fragments, colonies were picked and pooled to generate the library. Colony morphologies varied dramatically by size, colour, transparency and roughness, reflecting the effects of artificially increasing the copy number of numerous different genes. Over 97% of the colonies picked grew to turbid cultures following overnight growth in LB-Strep-Amp, indicating that overexpression of most genomic inserts did not adversely affect bacterial fitness.
Suppressor clones capable of growth in otherwise inhibitory concentrations of BBZ compounds were isolated on selective agar containing 1, 5, 10 or 20 times the MIC of BBZ5 and BBZ9. To confirm BBZ-resistance in these clones, suppressor colonies were isolated from selection plates and BBZ-resistance was confirmed by determination of MIC values for BBZ9. The identity of the genomic inserts in those plasmids that conferred BBZ resistance were determined by sequencing the 5ʹ and 3ʹ ends of the inserts using the primers M13-20 and M13Rev and comparing the obtained sequence with the annotated genome sequence of E. coli DH10B [20]. When this did not yield the complete insert, additional sequencing primers were designed to fill the gaps. Three unique inserts were identified in 12 sequenced inserts, all mapping to the same 6 kb stretch in the DH10B genome (Fig. 1). Only one ORF, that of the AraC family transcriptional regulator appY, was common to all three inserts.
Genomic inserts conferring resistance to BBZ compounds when present in high copy number. A Shown are the identity of sequenced inserts present in pUC19 that were isolated from BBZ-resistant suppressor strains. Large arrows represent open reading frames (ORFs), shaded arrows indicate ORFs for which the annotation was confirmed using BLASTX and white arrows indicate pseudogenes. Small arrows indicate the binding positions and directions of sequencing primers. B Genomic context of suppressor inserts within the DH10B genome (Durfee et al., 2008) and identities of the predicted protein products. Numbers are genomic co-ordinates, figures are to scale
Three additional independent screens of the genomic library for BBZ resistance were performed. In total, 69 plasmids conferring resistance to BBZ5 and BBZ9 were isolated. All of these were screened for the presence of appY by PCR using primers M13F_AD, appYF and appYR and were shown to contain the gene. To confirm that appY over-expression was the cause of BBZ-resistance, the native appY gene was cloned into the cis-repressed, IPTG-inducible vector pQE-80L. E. coli MC1061 was transformed with either pQE-80L or pQE-80L_appY and MIC assays were performed for BBZ4, BBZ5 and BBZ9 in the presence of 0, 0.1, 0.25 and 0.5 mM IPTG (Fig. 2). For MC1061 harbouring pQE-80L_appY, MIC values for all three compounds increased with IPTG concentration. IPTG did not significantly affect MIC values for the pQE-80L control. Similarly, the presence of the appY gene in the absence of IPTG-induced expression did not alter susceptibility of the bacteria to BBZ compounds. The up-regulation of appY expression from pQE-80L_appY was confirmed by qRT-PCR (Fig. 3), which showed that appY over-expression was responsible for the BBZ-resistance of the suppressor strains.
Over-expression of appY confers resistance to BBZ antibacterial compounds. A IPTG-induced expression of appY increases the MIC values of E. coli MC1061 to BBZ compounds. MICs of BBZ4, BBZ5, BBZ9 and ridinilazole were determined for E. coli MC1061 harbouring either pQE-80L (light grey bars) or pQE-80L_appY (dark grey bars), in the presence of increasing concentrations of IPTG. Data shown are the means ± SD of at least 3 independent experiments. B Confirmation of AppY-mediated BBZ resistance on solid media. E. coli MC1061 (1 × 107 CFU) harbouring pQE-80L (P), pQE-80L_appY (A) or pQE-80L_NHis6appY (N) was grown on LB agar containing 0x, 1x, 2x, 4x, 8x, 16x, 32x, and 64x the MIC (0.06 µg/ml) of BBZ5, BBZ9 or ridinilazole either in the absence or presence of 1 mM IPTG for 16 h at 37 °C. Induction of native and His6-tagged AppY by IPTG resulted in more than 64-fold suppression of the growth inhibitory activity of BBZ5, BBZ9 or ridinilazole. Scale bar, 0.5 cm
Measurement of appY expression from pQE-80L_appY by qRT-PCR. E. coli MC1061/pQE-80L_appY was induced with 1 mM IPTG. Shown is the relative abundance of the appY transcript for induced compared to un-induced cultures at each time point (grey bars), for un-induced cultures relative to t0 (□), and for induced cultures relative to t0 (△). All values were normalised to the abundance of 16 S rRNA and were significantly different from the comparison value (Students t test, p < 0.05) except the value marked * (p = 0.085)
AppY-mediated BBZ resistance does not proceed through AppY or known targets of AppY
There are several possible scenarios whereby AppY over-expression could confer resistance to BBZ antibacterials. On possibility is that AppY is itself the crucial molecular target. To assess the importance of AppY for bacterial growth, the expression plasmid pQE-80L_appY was used to transform E. coli strains MC1061 and the appY knockout JW0553. For both strains, growth was assessed with and without IPTG induction of appY expression (Supplementary Fig. S3). Deletion of appY did not affect viability or growth, as displayed by the normal growth kinetics of JW0553. Induction of the gene in this strain did however result in a modest inhibition of growth. Overall, AppY did not appear to be required for normal growth in complex media, or for BBZ susceptibility, indicating that BBZ-mediated growth inhibition does not proceed directly through AppY.
We next hypothesised that AppY could up-regulate the molecular target of the BBZ compounds. To assess if either of the three gene sets known to be induced by appY were indirectly responsible for the appY-mediated BBZ resistance, each of appA, appCB and hyaAF were cloned into pQE-80L. MIC values for BBZ5 and BBZ9 against MC1061 strains transformed with each vector were identical to wild type, with or without IPTG-induced transcription of the operons.
Identification of AppY-regulated genes using whole genome microarrays
The above results indicated that other, as-yet unidentified appY-regulated genes were responsible for BBZ resistance. To identify such genes, the appY regulon was determined using genome-wide transcriptional profiling. This method has been used to identify the complete regulon of transcriptional regulators SoxS and MarA in E. coli [21]. In this work, strains that carried chromosomal deletions for the regulator of interest were transformed with expression plasmids harbouring the relevant gene. This enabled tightly controlled user-defined expression of the two regulators. This approach was repeated here by exposing early-exponential phase cultures of JW0553/pQE-80L_appY to IPTG for 45 minutes.
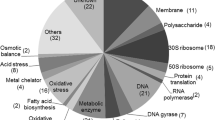
The induction of appY led to increased expression of 90 genes greater than 2.5-fold compared to the untreated control (Supplementary Table S1). Amongst these genes, as expected, were the appY-regulated genes appA, appC-B and hyaA-F as well as appY itself. Strikingly, 47 of the 90 up-regulated genes have defined or predicted roles in several stress responses (Table 2). Of these, 16, including the 6 most strongly up-regulated genes, were components of acid resistance (AR) systems AR2 and AR3 [22], and three transcriptional regulators including the central regulator of AR2, gadE, indicating that AppY may play a role as a regulator of acid resistance. Another 11 appY-induced genes are predicted to play roles in redox homoeostasis and protection from oxidative stress, and a further 20 have roles in diverse stress responses including osmotic stress, response to toxins and antibiotic resistance. In addition to these were a further 11 genes with metabolic functions and 16 genes whose functions were poorly defined. These observations indicated that appY-mediated BBZ resistance was an indirect effect of the induction of one or several stress responses.
AppY expression resulted in decreased abundance of 48 genes greater than 2.5-fold relative to the untreated control (Supplementary Table S2). All but 5 of these genes were arranged in 13 operons, indicating repression from relatively few promoters. The specific appY binding site has not been identified, and there is no evidence as yet that appY functions as a transcriptional repressor. The appY-repressed genes play roles in amino acid catabolysis, anaerobic respiration and carbohydrate metabolism, consistent with AppY’s predicted function as a coordinator of metabolic reprogramming under conditions of anaerobiosis and stationary phase growth [23].
Over expression of mdtEF confers resistance to BBZ compounds
To complement the initial MCS screen, a second round of MCS analysis was performed using genomic libraries derived from the strains JW0553 and HN818. JW0553 carries a chromosomal deletion for appY and was used to identify non-appY resistance determinants. In addition, to avoid isolation of the acrAB efflux system the acrA knockout strain HN818 was used. Screening of each library for BBZ resistance was performed as described for the MC1061 genomic library. Again, four unique inserts were identified that conferred resistance to BBZ compounds (Supplementary Table S3). Each insert contained at least one gene predicted to function in multi-drug export including mdtE and mdtF that were previously shown to be up regulated by appY over-expression. The independent isolation of mdtEF in this screen provided strong proof that AppY-mediated resistance to the BBZ compounds proceeded through up-regulation of the MdtEF-TolC efflux transporter.
E. coli lacking AppY shows impaired responses to acid and oxidative stresses
The large number of stress resistance genes up regulated by appY over-expression suggested that this gene may play an important role in instigating or regulating acid, oxidative and antibiotic stresses. To confirm the importance of AppY to these stress responses we assessed bacterial survival following acid shock in acidified media and survival after oxidative stress induced by hydrogen peroxide exposure (Fig. 4).
E. coli lacking AppY show markedly increased sensitivity to acid and oxidative stresses. A Schematic diagram of acid and oxidative resistance assays. Stationary phase cultures grown in the appropriate media were diluted 1:1000 into the test indicated challenge media. Viable counts were measured following serial dilution in PBS and plating on LB agar. Survival is given as a percentage of the viable counts taken at inoculation into the challenge media. Results of acid resistance assays as percentage cell survival for MG1655 (WT), JW0553 (ΔappY) and JW0553 complimented with an appY expression vector (ΔappY /pQE-80L_appY) with no pre-induction of AR systems (B), induction of AR1 (C), induction of AR2 (D) and induction of AR3 (E) following 2 h (white bars) or 4 h (red bars) challenge in acidified media. F. E. coli lacking AppY show increased sensitivity to oxidative stress. Graph shows percentage survival for the same three strains as above following 1 h in LB supplemented with 0% H2O2 (white bars), 0.001% H2O2 (light blue bars) and 0.01% H2O2 (dark blue bars). For all assays error bars indicate standard deviation for four biological replicates. Results are representative of at least 2 experimental repeats
The appY-induced transcription of multiple genes involved with multi-drug resistance, including the multi-drug efflux transporter MdtEF:TolC, has indicated that appY over-expression might confer non-specific resistance to several antibiotics. To assess this, MIC assays in MC1016/pQE-80L_appY with and without IPTG induced over-expression of AppY were repeated with ciprofloxacin, novobiocin, tetracycline and erythromycin. MIC values for erythromycin were increased at least 8-fold to ≥64 μg/ml, suggesting either overlapping MOAs or shared routes of cell entry or efflux for BBZs and erythromycin. No change in MIC was noted for the remaining antibiotics. AppY thus appears to play a role in resistance to a limited range of un-related antibacterial agents.
To test the importance of AppY in the response to acid stress, the survival of the appY wild type strain MG1655 (WT), the appY deletion strain JW0553 (ΔappY) and the appY deletion strain complemented with a native appY expression vector (ΔappY/pQE-80L_appY) was assessed after 2 h and 4 h exposure to acidified minimal media (pH ≤ 2). Cells lacking AppY were completely killed following 2 h acid treatment whilst complementation of the mutant strain restored wild type survival levels (Fig. 4b).
E. coli utilizes three AR systems to enable survival at low pH conditions [24]. AR1 is glucose-dependent and remains poorly characterized whereas AR2 and AR3 are well understood and can be induced by growth in defined media containing glutamate and arginine respectively [24]. To determine if AppY was involved in the induction of any specific AR system, cell survival following acid challenge was also determined with and without pre-induction of AR1, AR2 and AR3 (Fig. 4). Pre-induction of all three AR systems increased survival significantly, with induction of AR2 increasing cell survival to almost 100% even in the appY knockout strain. JW0553 survival was still impaired even with pre-induction of AR1 and AR3, and this was especially pronounced with AR1 pre-induction suggesting that AppY may be involved in the induction of this system. However, the increased survival with AR system pre-induction, even in strain JW0553, suggests that AppY induction of AR can be circumvented by other regulators when AppY is absent. Overall, this data indicates that AppY plays a key role in the early induction of acid resistance responses.
Since several genes induced by AppY have defined or putative roles in oxidative stress and/or redox homoeostasis, we also tested whether the appY deletion strain was hypersensitive to killing by hydrogen peroxide (Fig. 4f). Again, strain JW0553 showed a pronounced sensitivity to this extracellular stress compared to the wild type MG1655, and near wild type survival was restored by complementation with in-trans appY. The lack of AppY-mediated up-regulation of ahpC, a key component of the key E. coli hydrogen peroxide scavenger alkyl hydroperoxide reductase is a likely contributor to this effect [25].
Discussion
We report here that incremental increases of AppY expression from pQE-80L_appY resulted in increasing resistance to BBZ compounds, confirming the protein as a BBZ resistance determinant in E. coli. AppY is a transcriptional regulator of the AraC family that has been proposed to be a global regulator of energy metabolism genes in E. coli [15,16,17]. Expression of appY is increased under conditions of anaerobiosis, phosphate and carbon starvation and entry into stationary phase [26] and is repressed by the histone-like protein HNS through binding its promoter region in response to growth conditions such as cold-shock [27]. More recently appY has been identified in a genomic screen for genes that stabilised the stationary phase sigma factor, σS [28], itself a master transcriptional regulator of the metabolic changes associated with entry to stationary phase and the general stress response [29]. Thus, appY plays a role in integrating numerous signals regarding the metabolic state of the cell and bringing about appropriate cellular responses.
The cause of AppY-mediated BBZ resistance
Inhibition of the DNA-binding activities of AraC family transcription factors has been documented for 1-hydroxybenzimidazoles that have structures bearing some similarities to the BBZ molecules studied here [30]. The 1-hydroxybenzimidazoles have no intrinsic antibacterial activity. Instead, they were designed to mitigate the toxic effects of bacterial infections by inactivating virulence regulators such as SoxS and MarA. In addition to itself, appY over-expression increased the abundance of genes encoding the AraC family transcriptional regulators GadX and GadW. Therefore, if the BBZ compounds possess binding affinities for these regulators they could conceivably titrate the drugs away from their antibacterial target(s), explaining the observed increase in MIC values.
An alternative explanation for appY-mediated BBZ resistance is the induction of multiple stress response genes. The co-ordinated activation of acid, oxidative and general stress responses could prime the bacterium for the challenges brought about by BBZ treatment. Equally plausible is that one set of genes within this complex response network mediates resistance. The gene most strongly expressed following appY transcription was slp, which encodes the starvation lipoprotein. This gene, along with the co-transcribed dctR, greatly increases survival at acidic pH [31]. However, null mutants for slp do not show significant changes in resistance to chloramphenicol, peroxide, SDS, osmotic or temperature stress [32, 33], indicating that slp expression does not affect the permeability of the Gram-negative outer membrane. More likely to be of importance are MdtE and MdtF (formerly YhiU and YhiV), which form a multi-drug transporter with TolC [34] and have been implicated in resistance to several antibiotics including erythromycin [35, 36]. This is therefore the most likely cause of appY-mediated erythromycin resistance and the induction of mdtE and mdtF could similarly result in increased efflux of the BBZ compounds.
The contributions of other transcriptional regulators to the AppY regulon and implications for the physiological functions of AppY
Transcriptional profiling has been used to identify the regulons of a multitude of transcriptional regulators including SoxS, MarA, HNS, GadX, GadW and LuxS [21, 22, 37, 38]. The study performed by Pomposiello and colleagues, which determined the regulons of SoxS and MarA, was used as a blueprint for the experiments to identify the AppY regulon described in this chapter. For SoxS and MarA, transcriptional activity is solely regulated by intracellular concentration [21]. In this instance over-expression of a particular regulator in a strain lacking a chromosomal copy is expected to be sufficient to reveal the entire regulon using comparative transcriptional profiling. However, the activity of AppY is predicted to be modulated by an as-yet unidentified low-molecular-weight effector molecule produced under conditions of anaerobiosis and phosphate starvation [26]. The experimental set-up used here might therefore omit genes that are affected by AppY only under limiting oxygen or phosphate, or in the presence of the unknown effector. Nevertheless, the microarray analysis revealed 137 genes that were up-regulated and 81 genes that were down-regulated greater than 2.5-fold (P < 0.05) 45 min after appY induction.
The abundance of 9 transcriptional regulators was increased following appY induction indicating that AppY may influence several regulatory networks, and that these perturbations may have contributed to the gene lists. To assess these contributions each gene altered by appY was searched in the E. coli K-12 MG1655 database [39] for known regulators (Table 2).
The autoinducer-2 synthase, luxS, was strongly induced by appY. However, only two of the genes up-regulated greater than 2-fold by AppY (hyaC and sufS) were also in the previously described LuxS regulon [38, 40, 41]. Two less strongly induced genes, ihfA and ihfB, may have contributed to the up-regulation of six and down-regulation of two genes. IhfA and IhfB form the integration host factor heterodimer, a histone-like DNA binding complex that affects the expression of several genes, but which is more often associated with chromosome compaction [39, 42]. The contribution of other regulators appears more significant when the appY-induced acid resistance genes are considered. Of these, 11 out of 16 have previously been placed in the GadX-GadW regulon [22], both of which were up-regulated by AppY. In addition, the gene for GadE, which is essential for expression of the glutamate-dependant acid resistance system [43], was also strongly induced following appY over-expression. No connection has previously been observed between AppY and the regulatory networks controlling acid resistance. This data indicates that the regulator may play a role in mediating the physiological response to low pH, and this premise is supported by the increased susceptibility of the appY knockout strain to acid stress, oxidative stress and selected antibacterials demonstrated in this work.
The alternative sigma factor, σS (RpoS) is the central regulator controlling genes that mediate entry into stationary phase and control general stress responses [29]. Over-production of AppY stabilises σS [18], indicating that this activity could account for the general stress response genes observed in Table 2. However, only 11 of 90 AppY-induced genes have previously been shown to require RpoS for induction [44]. RpoS is actively degraded by ClpXP during exponential growth, silencing the σS regulon. This degradation requires the adaptor protein RssB to be bound to RpoS, in order for it to be recognised by the protease. Several anti-adaptors prevent the presentation of RssB to ClpX in response to various stresses such as phosphate starvation (IraP), limiting magnesium (IraM) and DNA damage (IraM) [18]. AppY is predicted to induce the expression of an additional RssB anti-adaptor that has not been identified. There were 18 genes induced greater than 2.5-fold following IPTG treatment whose altered abundance can, thus far, only be ascribed to appY. These include the chaperonin coding genes groL, groS and dnaK, the PBP3 protease gene prc and several conserved proteins of unknown function. One of the conserved proteins in this list may represent the hypothesised anti-adaptor. The currently identified anti-adaptors share no structural similarity, therefore predictions of which gene encodes the anti-adaptor cannot be based on sequence homology and instead would require σS degradation inhibition assays using purified proteins. This was beyond the scope of the work presented here Table 3.
Acid resistance
The data suggests that AppY may play a role in early response to acid stress, but that this role is redundant with pre-exposure to conditions that induce AR2 and AR3.
DNA binding and potential relevance to C difficile mode of action
The original design concept for the BBZ compounds optimised their binding to the AT-rich minor groove sequences of duplex DNA. The AppY protein is a DNA-binding transcription factor and the appY gene, comprising 750 nucleotides, is very AT-rich, with 512 AT base pairs. Within the gene there are four AATT and four ATAT sites, all of which are high-affinity sites for BBZ compounds [1, 13], suggesting that they may be regulating appY expression via a feed-back loop arrangement. However the data presented here is more consistent with BBZ interference with AppY transcription factor function rather than direct binding to the gene. The C. difficile genome [45] and laboratory strain variants (for example, strain 630Δerm [46]) are also highly AT-rich with 71% AT content, consistent with the recent findings [13] that the BBZ compounds BBZ5 and ridinilazole could have genomic DNA minor groove binding as their primary mode of action. Exact analogues of appY in C. difficile have not yet been established but several transcriptional regulators of toxin production in this organism have been identified such as RstA [47] and Lrp [48]. The latter gene, lrp, also has high AT content (67%: https://clostridium.biocyc.org/).
Materials and methods
Chemicals
BBZ compounds were synthesised and purified as described previously [1, 3]. Ridinilazole was a gift from Summit Therapeutics. Ampicillin, ciprofloxacin, erythromycin, kanamycin, tetracycline and streptomycin and IPTG were purchased from Sigma (Poole, UK). Novobiocin was purchased from Merck (NJ, USA). Ciprofloxacin was dissolved in acidified water. Ampicillin was dissolved in 50% (v/v) ethanol. All other antibiotics as well as IPTG were dissolved in HPLC grade water, filtered through 0.45 µm pore-size filter units (Millex, Ireland). Antibiotics were stored at 4 °C and 1 M stock solutions of IPTG were stored at −20 °C, prior to use.
Bacterial strains, plasmids and culture conditions
Bacterial strains and plasmids used in this study are listed in Supplementary Table S4. E. coli were grown in LB-Lennox broth (10 g/l peptone, 5 g/l yeast extract, 5 g/l NaCl) with shaking (200 r.p.m, 37 °C) and maintained on LB agar. MC1061 and derivative strains were selected by supplementation of media with 50 µg/ml streptomycin (Str). Strain JW0553 was maintained on media containing 50 µg/ml kanamycin (Kan). For selection and maintenance of pUC19, pQE-80L and derivative plasmids, culture media were further supplemented with 100 µg/ml ampicillin (Amp). BBZ-agar plates were prepared individually by the addition of 20 ml molten LB agar to 20 μl BBZ at 1,000x the desired final concentration, with mixing by vortexing prior to pouring. For acid and oxidative stress assays strains were grown in either LB broth buffered with 100 mM morpholinepropanesulfonic acid (MOPS, pH 8.0) or brain heart infusion (BHI) medium buffered with morpholineethanesulfonic acid (MES; pH 5.0). Where indicated these media were supplemented with 0.4% glucose (LBG and BHIG). The acid resistance assay medium was minimal EG adjusted to the required pH values with HCl.
Topoisomerase inhibition assays
Assessment of BBZ mediated inhibition of type II topoisomerases was performed as described previously [6] using purified E. coli enzymes and materials purchased from Inspiralis Ltd, (Norwich, UK).
Determination of MIC values
Minimal inhibitory concentrations (MIC) were determined using the broth dilution method [49] using quadruplicate increasing twofold dilutions of antibiotics in LB media. For assessment of the effects of gene induction on MIC values IPTG was included in the assay media at final concentrations 0.1 mM, 0.25 mM and 0.5 mM.
Scanning electron microscopy
DMSO or BBZ compounds at various multiples of the MIC were added to E. coli MC1061cultures immediately after inoculation. Cells were incubated for 8 h or 24 h, collected and washed three times in sterile PBS. Fixation, dehydration, mounting and gold coating and imaging were performed as described previously [6].
DNA manipulations and genetic constructions
All molecular manipulations were performed by standard methods unless stated otherwise [50]. Restriction endonucleases, Antarctic Phosphatase, T4 DNA ligase and standard PCR reagents were obtained from New England Biolabs (MA, USA). Ligations were performed using T4 DNA ligase (16 °C, 16 h), with a 3:1 molar ratio of insert:vector. Plasmids were isolated from bacterial strains using plasmid Quicklyse miniprep kits (Qiagen) following the manufacturer’s recommendations. Nucleic acid preparations were quantified and checked for purity using a Nanodrop ND-1000 spectrophotometer (Thermo Fisher Scientific). Transformations were performed using the heat-shock method with recovery in SOC media (2% w/v tryptone, 0.5% w/v yeast extract, 10 mM NaCl, 2.5 mM KCl, 10 mM MgCl2, 20 mM MgSO4, 20 mM glucose) for 1 h at 37 °C followed by plating on LB-Str-Amp agar (37 °C, 18 h). Oligonucleotide primers were designed using Primer3Plus [51] and purchased from Invitrogen. Primer sequences are listed in Supplementary Table S5. DNA sequencing was performed at the DNA Sequencing Facility, Wolfson Institute for Biomedical Research, University College London. Nucleotide sequence data were analysed using MacSequenceView and compared with sequences in the NCBI genomes database using BLAST [52].
Construction of E. coli genomic libraries
E. coli genomic libraries were constructed as 3–4 kb Sau3AI genomic fragments cloned into the BamHI site of pUC19 using the following procedure. Genomic DNA was isolated from stationary phase cultures of E. coli MC1061, JW0553 and HN818 using the DNeasy Blood and Tissue kit (Qiagen). Following partial digestion of gDNA with Sau3AI (0.25 U, 37 °C, 1 h), 3–4 kb fragments were isolated by extraction from 1% agarose gels and purified on Qiaquick columns (Qiagen). The pUC19 vector was cleaved with BamHI and treated with Antarctic Phosphatase to prevent self-ligation. Genomic fragments were then ligated into the pUC19 BamHI site. E. coli MC1061 was made chemically competent by the Inoue method [53]. The pUC19:3–4 kb gDNA ligation mix was used directly to transform MC1061. Transformants were recovered in SOB broth (SOC media without 20 mM glucose) for 1 h at 37 °C and plated on SOB-Str-Amp agar. Following overnight growth at 37 °C, colonies were transferred using sterile toothpicks to single wells in 96-well plates containing 200 µl LB-Str-Amp broth. Each plate contained control wells to confirm LB sterility, positive growth of un-transformed MC1061 in LB-Strep and negative growth in LB-Strep-Amp. Following overnight growth in 96-well plates (37 °C, 18 h, 200 r.p.m.), 50 μl of culture from wells in which colonies grew were pooled, mixed by vortexing, combined with an equal volume of 30% glycerol and stored as aliquots at −80 °C.
For an average genomic insert size of 3.5 kb and given a genomic size of 4.64 Mb for MC1061, a library of 10,000 clones was sufficient to include every possible fragment once with a probability of 99.9%. For each strain a total of 15 sub-libraries, were constructed on separate days. For each strain in turn these libraries were pooled, generating the selection pools which comprised >10,000 individually selected transformants. In addition, separate libraries of >20,000 MC1061, JW0553 or HN818 clones transformed with pUC19 were constructed as control pools.
Construction of expression plasmids
To generate a construct that enabled expression of native, untagged AppY the appY gene from E. coli MC1061 was amplified by PCR using the primers EcoRI_rbs_appY_F and PstI_appY_R, which contain the indicated restriction sites (Supplementary Table S5). In addition, EcoRI_RBS_appY_F contains the consensus ribosome-binding site (RBS, AAAGAGGAG) downstream of the EcoRI site. The amplified DNA containing the appY gene was purified by gel extraction and cut with EcoRI and PstI. In parallel the cis-repressed expression vector pQE-80L (Qiagen) was digested with the same enzymes removing the vector’s His6 tag. The appY fragment was then ligated into the open vector and the ligation mix was used to transform E. coli MC1061. Positive recombinants were screened by restriction mapping of plasmids isolated from clones that grew on selective plates. Positive candidates were confirmed by sequencing using the primers pQEF and pQER. The final construct maintained the appY insert with 100% sequence identity to the appY gene in E. coli DH10B and incorporated the consensus RBS within EcoRI_RBS_appY_F at the -10 position relative to the ATG start codon. This construction was named pQE-80L_appY and was used to transform “wild type” E. coli MC1061, yielding the clone MC1061/pQE-80L_appY. This clone was used in all subsequent appY induction MIC assays.
Plasmids pQE-80L_appA and pQE-80L_appC-B were constructed in the same manner as pQE-80L_appY using the primers indicated in Supplementary Table S5. For inducible expression of the hyaA-F operon, pQE-80L_hyaAF was constructed. The coding region of all six genes was amplified as a single construct using the primers indicated in Supplementary Table S5 and the LongRange PCR kit (Qiagen). The cloning procedure was otherwise identical to that of pQE-80L_appY. The resulting plasmids contained a T5 promoter under the control of LacI, enabling IPTG-driven transcription of the encoded sequences. All sequences included the -10 and -35 sequences upstream of the first gene in the operon, thus maintaining the operonic ribosome-binding site. Expressed proteins were native, as the digestion of pQE-80L with EcoRI and PstI removed the vector’s His6 tag. All inserts were confirmed by restriction mapping of the resulting expression vectors, producing bands of the expected size in each case.
Confirmation of appY expression from pQE-80L_appY
Expression of appY from pQE-80L_appY was confirmed using two-step qRTPCR. Overnight cultures of E. coli MC1061, freshly transformed with pQE-80L_appY, were used to inoculate six cultures of 50 ml LB-Str-Amp to a starting OD595 of 0.05. At logarithmic growth phase (OD595 ~1.0) triplicate cultures were either left untreated or treated with IPTG at a final concentration of 1 mM to induce appY expression. Bacterial cells from induced or un-induced cultures were pooled and harvested at 0, 1, 4 and 22.5 h post-induction by centrifugation (5 min, 13,000 r.p.m). The abundance of AppY was Total RNA was stabilised and isolated and converted to cDNA and qPCR was performed as described previously [6]. Primer sequences for qPCR were designed using Primer3Plus [51] and are given in Supplementary Table S5. Data was normalised to 16 S rRNA. Primers were used at concentrations of 300 nM (16 S rRNA) and 1200 nM (appY).
Transcriptional analysis
Overnight cultures of E. coli JW0553, freshly transformed with pQE-80L_appY, were used to inoculate 50 ml LB supplemented with Amp and Kan to a starting OD595 of 0.05. At early logarithmic growth phase (OD595 ~ 0.5), triplicate cultures were either left untreated or treated with IPTG at a final concentration of 1 mM to induce appY expression. Bacterial cells (2 ml) from induced or un-induced cultures were pooled and harvested 45 min post-induction by centrifugation (5 min, 13,000 r.p.m.). The mRNA profile at the time of harvesting was stabilised using RNAProtect Bacterial Reagent (Qiagen). Experiments were performed four times on separate days. Cell pellets were stored at −80 °C prior to RNA extraction. The preparation of total cellular RNA and conversion to Cy5-dCTP labelled cDNA was performed as described previously for S. aureus [6]. For normalisation, JW0553 genomic DNA was labelled with dUTP-Cy3 using the Genomic DNA Enzymatic Labelling Kit (Agilent). DNA and RNA were then co-hybridised onto E. coli K-12 8x15k format gene expression arrays [54] (Oxford Gene Technology, Oxford, UK). Hybridisation and washing of arrays were performed using the Oligo aCGH Hybridisation Kit and Oligo aCGH Wash Buffer Kit according to the manufacturer’s recommendations. Scanning and feature extraction was undertaken using an Agilent DNA microarray scanner and Agilent Feature Extraction Software v 10.7 respectively. Statistical analysis was performed using GeneSpring v11.1 software. Differentially expressed genes were defined as those that showed >2.5-fold up- or down-regulation compared to un-induced controls, with a P value of <0.05 determined by one-way analysis of variance (ANOVA) with Benjamini and Hochberg false discovery rate correction. Fully annotated microarray data has been deposited in BµG@Sbase and ArrayExpress and may be obtained from the St George’s facility.
Acid and oxidative stress resistance assays
Acid resistance (AR) assays were performed using the method of Richard and Foster [55]. Briefly, strains were grown overnight without pre-induction of AR (LB MOPS pH 8.0), with pre-induction of AR1 (LB MES pH 5.5), with pre-induction of AR2 (LBG pH 5.5) or with pre-induction of AR3 (BHIG pH 5.5). Quadruplicate stationary-phase cultures were normalised to the lowest OD (600 nm) of all strains, diluted 1:1,000 into pre-warmed pH 2.5 EG medium without amino acid supplementation (for control and AR1), with 1.6 mM glutamate (for AR2), or with 1.0 mM arginine (AR3). Oxidative stress assays were performed in the same way as the acid resistance assays using overnight cultures grown in LB (MOPS pH 8) diluted 1:1000 into LB supplemented with increasing concentrations of hydrogen peroxide. Following 1 h, 2 h and 4 h incubations, cells were enumerated by serial dilution in PBS, followed by growth (24 h, 37 °C) and colony counting on LB plates. Viable counts (CFU/ml) were determined, and percent survival was calculated relative to time zero for each strain under each condition.
References
Neidle S, Mann J, Rayner EL, Baron A, Opoku-Boachen Y, Simpson IJ, et al. Symmetric bis-benzimidazoles, a new class of sequence-selective DNA-binding molecules. J Chem Soc Chem Commun. 1999;929–30..
Bailly C, Chessari G, Carrasco C, Joubert A, Mann J, Wilson WD, et al. Sequence-specific minor groove binding by bis-benzimidazoles: water molecules in ligand recognition. Nucleic Acids Res. 2003;31:1514–24.
Mann J, Baron A, Opoku‐Boahen Y, Johansson E, Parkinson G, Kelland LR, et al. A new class of symmetric bisbenzimidazole‐based DNA minor groove‐binding agents showing antitumor activity. J Med Chem. 2001;44:138–44.
Seaton A, Higgins C, Mann J, Baron A, Bailly C, Neidle S, et al. Mechanistic and anti-proliferative studies of two novel, biologically active bis-benzimidazoles. Eur J Cancer. 2003;39:2548–55.
Bis-benzazoles and their use as antineoplastic agents. Neidle S, Mann J, 2000, WO 00/63180
Dale AG, Hinds J, Mann J, Taylor PW, Neidle S. Symmetric bis-benzimidazoles are potent anti-staphylococcal agents with dual inhibitory mechanisms against DNA gyrase. Biochemistry. 2012;51:5860–71.
Moreira JB, Mann J, Neidle S, McHugh TD, Taylor PW. Antibacterial activity of head-to-head bis-benzimidazoles. Int J Antimicrob Agents. 2013;42:361–6.
Mann J, Taylor PW, Dorgan CR, Johnson PD, Wilson FX, Vickers R, et al. The discovery of a novel antibiotic for the treatment of Clostridium difficile infections: a story of an effective academic-industrial partnership. Medchemcomm. 2015;6:1420–6.
Cho JC, Crotty MP, Pardo J. Ridinilazole: a novel antimicrobial for Clostridium difficile infection. Ann Gastroenterol. 2019;32:134–40.
Thorpe CM, Kane AV, Chang J, Tai A, Vickers RJ, Snydman DR. Enhanced preservation of the human intestinal microbiota by ridinilazole, a novel Clostridium difficile-targeting antibacterial, compared to vancomycin. PLoS One. 2018;13:e0199810.
Snydman DR, McDermott LA, Thorpe CM, Chang J, Wick J, Walk ST, et al. Antimicrobial susceptibility and ribotypes of Clostridium difficile isolates from a Phase 2 clinical trial of ridinilazole (SMT19969) and vancomycin. J Antimicrob Chemother. 2018;73:2078–84.
Vickers RJ, Tillotson GS, Nathan R, Hazan S, Pullman J, Lucasti C, et al. CoDIFy study group. Efficacy and safety of ridinilazole compared with vancomycin for the treatment of Clostridium difficile infection: a phase 2, randomised, double-blind, active-controlled, non-inferiority study. Lancet Infect Dis. 2017;17:735–44.
Mason C, Avis T, Hu C, Naqalingam N, Mudaliar M, Coward C, et al. The novel DNA binding mechanism of ridinilazole, a precision Clostridiodes difficile antibiotic. Research Square, 2022. https://doi.org/10.21203/rs.3.rs-1396940/v1
Li X, Zolli-Juran M, Cechetto JD, Daigle DM, Wright GD, Brown ED. Multicopy suppressors for novel antibacterial compounds reveal targets and drug efflux susceptibility. Chem Biol. 2004;11:1423–30.
Atlung T, Nielsen A, Hansen FG. Isolation, characterization, and nucleotide sequence of appY, a regulatory gene for growth-phase-dependent gene expression in Escherichia coli. J Bacteriol. 1989;171:1683–91.
Atlung T, Brøndsted L. Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation, and growth phase. J Bacteriol. 1994;176:5414–22.
Brøndsted L, Atlung T. Anaerobic regulation of the hydrogenase 1 (hya) operon of Escherichia coli. J Bacteriol. 1994;176:5423–8.
Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. Multiple pathways for regulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol. 2008;68:298–313.
Li X, Zolli-Juran M, Cechetto JD, Daigle DM, Wright GD, Brown ED. Multicopy suppressors for novel antibacterial compounds reveal targets and drug efflux susceptibility. Chem Biol. 2004;11:1423–30.
Durfee T, Nelson R, Baldwin S, Plunkett G 3rd, Burland V, Mau B, et al. The complete genome sequence of Escherichia coli DH10B: insights into the biology of a laboratory workhorse. J Bacteriol. 2008;190:2597–606.
Pomposiello PJ, Bennik MH, Demple B. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J Bacteriol. 2001;183:3890–902.
Tucker DL, Tucker N, Ma Z, Foster JW, Miranda RL, Cohen PS, et al. Genes of the GadX-GadW regulon in. Escherichia coli J Bacteriol. 2003;185:3190–201.
Brøndsted L, Atlung T. Effect of growth conditions on expression of the acid phosphatase (cyx-appA) operon and the appY gene, which encodes a transcriptional activator of Escherichia coli. J Bacteriol. 1996;178:1556–64.
Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2:898–907.
Seaver LC, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol. 2001;183:7173–81.
Atlung T, Knudsen K, Heerfordt L, Brøndsted L. Effects of sigmaS and the transcriptional activator AppY on induction of the Escherichia coli hya and cbdAB-appA operons in response to carbon and phosphate starvation. J Bacteriol. 1997;179:2141–6.
Atlung T, Sund S, Olesen K, Brøndsted L. The histone-like protein H-NS acts as a transcriptional repressor for expression of the anaerobic and growth phase activator AppY of Escherichia coli. J Bacteriol. 1996;178:3418–25.
Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S. Multiple pathways forregulation of sigmaS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol Microbiol. 2008;68:298–313.
Loewen PC, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–17.
Bowser TE, Bartlett VJ, Grier MC, Verma AK, Warchol T, Levy SB, et al. Novel anti-infection agents: small-molecule inhibitors of bacterial transcription factors. Bioorg Med Chem Lett. 2007;17:5652–5.
Mates AK, Sayed AK, Foster JW. Products of the Escherichia coli acid fitness island attenuate metabolite stress at extremely low pH and mediate a cell density-dependent acid resistance. J Bacteriol. 2007;189:2759–68.
Alexander DM, St John AC. Characterization of the carbon starvation-inducible and stationary phase-inducible gene slp encoding an outer membrane lipoprotein in Escherichia coli. Mol Microbiol. 1994;11:1059–71.
Price GP, St John AC. Purification and analysis of expression of the stationary phaseinducible slp lipoprotein in Escherichia coli: role of the Mar system. FEMS Microbiol Lett 2000. 2000;193:51–56.
Stenberg F, Chovanec P, Maslen SL, Robinson CV, Ilag LL, von Heijne G, et al. Protein complexes of the Escherichia coli cell envelope. J Biol Chem. 2005;280:34409–33419.
Nishino K, Yamaguchi A. Overexpression of the response regulator evgA of the two-component signal transduction system modulates multidrug resistance conferred by multidrug resistance transporters. J Bacteriol. 2001;183:1455–8.
Nishino K, Yamaguchi A. EvgA of the two-component signal transduction system modulates production of the yhiUV multidrug transporter in Escherichia coli. J Bacteriol. 2002;184:2319–21.
Hommais F, Krin E, Laurent-Winter C, Soutourina O, Malpertuy A, Le Caer JP, et al. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol Microbiol. 2001;40:20–36.
Wang L, Li J, March JC, Valdes JJ, Bentley WE. luxS-dependent gene regulation in Escherichia coli K-12 revealed by genomic expression profiling. J Bacteriol. 2005;187:8350–60.
Freundlich M, Ramani N, Mathew E, Sirko A, Tsui P. The role of integration host factor in gene expression in Escherichia coli. Mol Microbiol. 1992;6:2557–63.
Alekshun MN, Levy SB. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother. 1997;41:2067–75.
ECOCYC. Encyclopaedia of the Escherichia coli K-12 genes and metabolism. Website: www.ecocyc.org (Date last accessed 30 March 2022).
Sarkar T, Petrov AS, Vitko JR, Santai CT, Harvey SC, Mukerji I, et al. Integration host factor (IHF) dictates the structure of polyamine-DNA condensates: implications for the role of IHF in the compaction of bacterial chromatin. Biochemistry. 2009;48:667–75.
Ma Z, Gong S, Richard H, Tucker DL, Conway T, Foster JW. GadE (YhiE) activates glutamate decarboxylase-dependent acid resistance in Escherichia coli K-12. Mol Microbiol. 2003;49:1309–20.
Lacour S, Landini P. SigmaS-dependent gene expression at the onset of stationary phase in Escherichia coli: function of sigmaS-dependent genes and identification of their promoter sequences. J Bacteriol. 2004;186:7186–95.
Sebaihia M, Wren BW, Mullany P, Fairweather NF, Minton N, Stabler R, et al. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat Genet. 2006;38:779–86.
van Eijk E, Anvar SY, Browne HP, Leung WY, Frank J, Schmitz AM, et al. Complete genome sequence of the Clostridium difficile laboratory strain 630Δerm reveals differences from strain 630, including translocation of the mobile element CTn5. BMC Genomics. 2015;16:31.
Edwards AN, Anjuwon-Foster BR, McBride SM. RstA is a major regulator of clostridioides difficile toxin production and motility. mBio. 2019;10:e01991–18.
Chen KY, Rathod J, Chiu YC, Chen JW, Tsai PJ, Huang IH. The transcriptional regulator Lrp contributes to toxin expression, sporulation, and swimming motility in clostridium difficile. Front Cell Infect Microbiol. 2019;9:356.
Andrews JM. Determination of minimum inhibitory concentrations. J Antimicrob Chemother. 2001;48:5–16.
Sambrook J, Green MR Molecular Cloning: A Laboratory Manual (Fourth Edition). Cold Spring Harbor Laboratory Press, 2012.
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–74. https://primer3plus.com
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–402.
Inoue H, Nojima H, Okayama H. High efficiency transformation of Escherichia coli with plasmids. Gene. 1990;96:23–28.
Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–7.
Richard H, Foster JW. Escherichia coli glutamate- and arginine-dependent acid resistance systems increase internal pH and reverse transmembrane potential. J Bacteriol. 2004;186:6032–41.
Acknowledgements
Work at the School of Pharmacy was funded by The Wellcome Trust Seeding Drug Discovery Programme through their support of the drug discovery programme at Summit Therapeutics Ltd. A.G.D. was supported by a Medical Research Council Capacity Building Studentship. A.P. was supported by an Erasmus award. We thank the Wellcome Trust for funding BμG@S (the Bacterial Microarray Group at St. George’s) to provide microarray facilities and support. We are grateful to Jason Hinds at St George’s for help in using the facilities. Earlier work on the BBZ compounds was supported in part by Cancer Research UK. We are pleased to contribute this manuscript to the special issue of the journal honoring the contributions of Laurence Hurley. He has notably pioneered the application of chemical biology and chemical genetics to the study of the mode of action of nucleic acid binding drugs, especially involving higher-order DNA. He has influenced the direction of drug discovery in this field and his work has led directly to new agents in the clinic.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dale, A.G., Porcu, A., Mann, J. et al. The mechanism of resistance in Escherichia coli to ridinilazole and other antibacterial head-to-head bis-benzimidazole compounds. Med Chem Res 31, 1176–1191 (2022). https://doi.org/10.1007/s00044-022-02918-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00044-022-02918-7