Abstract
Background
Abundantly expressed factors in the oocyte cytoplasm can remarkably reprogram terminally differentiated germ cells or somatic cells into totipotent state within a short time. However, the mechanism of the different factors underlying the reprogramming process remains uncertain.
Methods
On the basis of Yamanaka factors OSKM induction method, MEF cells were induced and reprogrammed into iPSCs under conditions of the oocyte-derived factor Wdr82 overexpression and/or knockdown, so as to assess the reprogramming efficiency. Meanwhile, the cellular metabolism was monitored and evaluated during the reprogramming process. The plurpotency of the generated iPSCs was confirmed via pluripotent gene expression detection, embryoid body differentiation and chimeric mouse experiment.
Results
Here, we show that the oocyte-derived factor Wdr82 promotes the efficiency of MEF reprogramming into iPSCs to a greater degree than the Yamanaka factors OSKM. The Wdr82-expressing iPSC line showed pluripotency to differentiate and transmit genetic material to chimeric offsprings. In contrast, the knocking down of Wdr82 can significantly reduce the efficiency of somatic cell reprogramming. We further demonstrate that the significant suppression of oxidative phosphorylation in mitochondria underlies the molecular mechanism by which Wdr82 promotes the efficiency of somatic cell reprogramming. Our study suggests a link between mitochondrial energy metabolism remodeling and cell fate transition or stem cell function maintenance, which might shed light on the embryonic development and stem cell biology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mammalian oocyte cytoplasm includes abundant powerful reprogramming factors, which can remarkably reprogram terminally differentiated somatic cells into totipotent state within a short time. This reprogramming is the basic principle underlying somatic cell nuclear transfer (SCNT) and/or somatic cell cloning. Therefore, it has been previously speculated that oocyte-derived transcripts or proteins promote somatic cell reprogramming [35, 37]. It has also been suggested that Yamanaka factors (Oct4, Sox2, Klf4, c-Myc, collectively called OSKM factors) increase the speed and efficiency of the reprogramming process [33]. Induced pluripotent stem cells (iPSCs) have garnered considerable attention for use in patient-specific stem cell production and/or individualized treatment in mammalian animals, including scientists interested in their benefits for technical simplification and lack of ethics concerns. However, the current iPSC generation technology is still limited by bottlenecks, such as low induction efficiency, poor safety profile and long experimental period when compared with somatic cell nuclear transfer (SCNT) [17, 24]. To address the shortcomings of iPSC generation, the molecular mechanisms of SCNT have again attracted the interest of scientific researchers because of the similarity of cell fate reprogramming mediated by SCNT and iPS induction.
In fact, increasing evidence is revealing the molecular mechanisms underlying mouse embryonic fibroblast reprogramming, which is roughly categorized into initiation, maturation and stability stages. In the initiation stage of reprogramming, a wave of transcriptional and epigenetic modifications lead to changes in cell proliferation, metabolism and/or extracellular matrix remodeling. The acceleration of the cell cycle is conducive to reprogramming [14], for example, silencing of the tumor suppressor factor p53 accelerated reprogramming [18] due to its inhibitory effects on cell cycle progression. Zscan4 has been shown to prolong telomere life [23] and to act in conjunction with Yamanaka factors to inhibit the DNA damage response (DDR), stabilize genomic DNA and downregulate p53, thereby improving the efficiency of iPSC generation [16]. Regarding cellular metabolism reprogramming, Tcl1 inhibits the mitochondrial localization of polynucleotide phosphorylase (PnPase), suppressing mitochondrial biogenesis and oxidative phosphorylation and promoting energy metabolism remodeling, thereby promoting reprogramming [19]. Glis1 enables reprogramming of senescent cells into pluripotent cells by binding to chromatin at glycolytic genes, thereby enhancing acetylation (H3K27Ac) and lactylation (H3K18la) at pluripotency gene loci and facilitating cellular programming [26], indicating the link between somatic cell reprogrramming and cell fate determination [27, 28]. For extracellular matrix remodeling, Obox1, that is, oocyte-specific homeobox 1, facilitates cell reprogramming by promoting the mesenchymal-to-epithelial transition (MET) process and attenuating cell hyperproliferation [37]. Wdr5, Brca1 and Bard1 work together in response to DNA damage, and their absence affects the MET in the early stages of reprogramming [31].
The mature and stable reprogramming stage is based mainly on epigenetic modifications and pluripotency network maintenances. Dppa2 and Dppa4, key components of the chromatin-remodeling network, control pluripotency transformation by forming heterodimers, acting in conjunction with Yamanaka factors and remodeling chromatin configuration, thereby enhancing reprogramming [15]. The DNA methylation hydroxylase Tet1 cooperates with Oct4 in reprogramming and thus produce high-quality iPSCs [5]. Knockout of histone deacetylase 2 (HDAC2) promotes pluripotency factor binding to TET1, thereby reducing the DNA demethylation rate. Studies have shown that HDAC2-TET1 is a switch that regulates the maturation of pre-iPSCs into iPSCs [36]. Changes in epigenetic modifications affect the reprogramming stage. Zscan4f, a member of the ZSCAN protein family, recruits TET2. The Zscan4f-TET2 interaction promotes DNA demethylation and regulates the expression of downstream target genes associated with cellular metabolism and proteasome function, ultimately promoting iPSC generation [6].
In terms of epigenetic modification and pluripotency gene expression, WD repeat domain 82 (Wdr82) attracted our attention. First, Wdr82 has been identified as an oocyte-derived protein via mass spectrometry and RNA sequencing and is abundantly expressed in MII oocytes [12, 35], and the highly expressed oocyte cytokines had been reported to promote reprogramming. Second, the absence of Wdr82 reduces the expression level of pluripotent gene Oct4, hindering embryonic growth and development, and even leading to embryonic death [3]. On the other hand, the expression level of Wdr82 is significantly reduced during mESC differentiation, and the cell proliferation rate is significantly slowed after Wdr82 knockdown, indicating that Wdr82 plays an important role in mESC proliferation and differentiation in vitro [3, 42]. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A/B histone H3K4 methyltransferase complex and RNA polymerase II. The Wd82 component of the complex interacts not only with the RNA recognition motif of Setd1A but also the phosphorylated C-terminal domain of RNA polymerase II, thereby recruiting the Setd1A/B complex to the transcription starting sites of certain genes and catalyzing the histone H3K4 trimethylation of adjacent nucleosomes, activating their transcription [25, 38]. The loss of Wdr82 specificity resulted in a significant reduction of H3K4 trimethylation level of transcriptional genes, such as Oct4 [3, 38]. Hence, whether the highly expressed oocyte factors can increase the reprogramming efficiency of mouse somatic cells and the mechanism of Wdr82 function in iPSC generation become the foci of our attention. Here, we show that the overexpression of Wdr82, as the basis of OSKM factor-mediated reprogramming, significantly promotes the generation of iPSCs. Further analysis indicates that the overexpression of Wdr82 affects the cellular metabolic pathway by inhibiting oxidative phosphorylation in mitochondria during the early stage of somatic cell reprogramming.
Materials and methods
Ethical statement
All animal experiments were carried out according to the Regulations for the Administration of Affairs Concerning Experimental Animals published by the Ministry of Science and Technology, China (2004). All animal maintenance and experimental procedures were performed according to the Tongji University Guide for the use of laboratory animals.
Contact for reagent and resource sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Shaorong Gao (gaoshaorong@tongji.edu.cn).
Animals
ICR pregnant mice were purchased from the Beijing Sibeifu Company. C57BL/6 mice, OSKMOR transgenic mice and Oct4−GFP mice were purchased or otherwise obtained from Beijing Weitong Lihua Company, Shanghai Slack Laboratory Animal Co., Ltd., and the Jackson Laboratory, respectively. All mice involved in the study were SPF (specific pathogen-free) animals.
Plasmids
A pFuw-TetOn retroviral vector was used as the backbone to construct plasmids expressing Oct4, Sox2, Klf4, c-Myc, Wdr82, Dpy30 or Taf7. The primer pairs used to prepare constructs are listed in Table S1. Wdr82-shRNA were cloned into a psiccoR-RFP retroviral vector. The shRNA sequences are listed in Table S2.
Reprogramming to transition MEFs into iPSCs
One day before infection, OR-MEF/OG2-MEF cells were resuscitated and inoculated at 2.0 × 104 cells/well into a 12-well plate. The cells were then transfected with HEK293 cell-produced viral fragments carrying fusions of Oct4, Sox2, Klf4 and c-Myc genes (named OSKM factors) and/or Wdr82, with 5 μg/mL polybrene. After infection for 10–12 h, the culture medium was replaced to ESM containing Dox, and reprogramming induction was initiated, which was recorded as Day 0. Then, the culture medium was changed using replaced with fresh ESM medium supplemented Dox the next day, depending on the colony situation. Dox was removed until a large number of Oct4-GFP fluorescence-emitting clones were observed. Subsequent experiments, including flow cytometry and AP staining, and the establishment of the OSKM + Wdr82 iPSC line realized by selecting monoclonal cells, were performed when a Oct4-GFP-positive clone culture remained stable for three days. During the reprogramming process, the colony number was recorded every day, and Oct4-GFP-positive clones were also observed and imaged with a fluorescence microscope (IX73, Olympus, Japan).
RT-PCR
A PCR system was prepared on ice, and each sample was prepared as three biological replicates. Every PCR was performed using 2 × RealStar Fast SYBR qPCR Mix (HIGH ROX) (GenStar, A303-05) with primers and cDNA (5 ng). The reactions were run in triplicate on a fluorescence quantitative detection system (Bio–Rad) following the manufacturer’s instructions. Cycle threshold values were normalized to murine housekeeping gene Gapdh or Hprt mRNA expression. The primer pairs used for assessing the expression of exogenously added and/or endogenously expressed Oct4, Sox2, Klf4, c-Myc and other pluripotency genes are listed in Tables S3 and S4.
Flow cytometry
After the reprogramming induction period was completed, the original medium was removed by aspiration, and the cells were washed 1–2 times with DPBS; 300 μL of trypsin was used for digestion; and the same amount of FM medium was added to stop the trypsin digestion; and then, the cells were centrifuged at 1000 rpm at room temperature for 5 min. Next, 1 mL of the cell sample resuspended in DPBS was obtained, and computer-based analysis and detection of Oct4-GFP-positive and Oct4-GFP-negative cells were identified according to the BD LSRFortessa flow cytometry-based computer operation manual.
Alkaline phosphatase (AP) staining
After the completion of reprogramming induction, well-conditioned iPSC clones were selected for AP staining. The clones were washed 3 times with DPBS, and then 4% paraformaldehyde was added to fix the cells for 5 min or overnight at 4 °C. Then, the fixative was discarded, and the cells were washed 3 times with DPBS. A staining solution was then added to the cells in the dark, followed by washing with DPBS. The AP staining results were imaged, and the number of AP-positive clones was counted.
Embryoid body differentiation experiment
The hanging drop method was used to perform a EB differentiation experiment. The confluent iPSCs were trypsinized into single cells, followed by the removal of feeder cells. Then, the cell density was determined and adjusted to 5 × 104/mL in ESM (without LIF) after gentle pipetting. The cell suspension was evenly arranged on the lid of a 10-cm cell culture dish at a concentration of 20 μL cell suspension/drop; after 48 h of incubation, the formation of EB spheres in the hanging drop was observed. The EBs were collected and treated with 0.1% gelatin in a six-well plate, with the medium was changed every other day. Then, the cells were cultured in a 37 ℃ incubator for 3 days (the EB spheres were allowed to adhere to the plate walls and differentiate). At the end of the differentiation period, total RNA was extracted from the differentiated cells and reverse transcribed to evaluate germ layer marker gene expression. The primer pairs used for the assessment of the marker genes in the differentiated germ layers are listed in Table S5.
Immunofluorescence staining
The iPSC line was cultivated on a cell slide in a 24-well plate until the clone reached the proper density and size had grown to a suitable size. Then, the ESM medium was aspirated, and the cells were washed with DPBS, fixed at room temperature for 2 h, permeabilized with 0.1% Triton X-100 solution for 15 min at room temperature, blocked with a 3% BSA solution for 1 h at room temperature, incubated with primary antibody and secondary antibody for 1 h, and incubated with DAPI nuclear-staining solution for 5–10 min. Then, antifluorescence quenching solution was dropped on each slide. Each cell slide was placed face down, onto a glass slide, and finally sealed with nail polish. The slides were scanned and imaged with a Leica TCS SPE confocal microscope.
The following primary antibodies were used: anti-SSEA1 (Abcam, ab16285), anti-NANOG (CST, #8822S), anti-SOX2 (Abcam, ab97959) and anti-H3K4me3 (Bioss, bsm-33110 M). The following secondary antibodies were used: goat antimouse IgG Alexa Fluor 594 (Life, A-11005) and donkey antirabbit IgG Alexa Fluor 594 (Life, A-21207).
Karyotype analysis
The cultured iPS cell lines (OSKM + Wdr82 and/or OSKM) are removed the original culture medium and add culture medium containing 0.25 g/mL colchicine (sigma, C-9754), incubating for 4–8 h until most clones show grape-like shape, being arrested in the metaphase. Then the colonies are digested into single cells by using trypsin at 37 ℃, followed by the trypsin neutralization and centrifuge at 1000 rpm for 5 min. Discard the supernatant, and resuspend the cells in 1 mL of preheated hypotonic solution (0.4 M sodium citrate and 0.4 M KCl in a 1:1 ratio) and then incubate in a 37 ℃ water bath for 5 min. Then the cells are prefixed and fixed by adding seven drops and 4 mL of fixation solution (methanol: glacial acetic acid in a 3:1 ratio), respectively, for 40 min at room temperature. After the second fixation, centrifuge at 1000 rpm for 5 min, discard the supernatant, and add 50 μ L fixation solution, gently resuspend cell precipitation using a Babbitt tube. The cell suspension are dropped onto the precooled glass slides from a height of approximately 1.5 m. Place the glass slides in a 65 ℃ oven for more than 2 h, followed by Giemsa staining (dilute 10 times of Giemsa concentrate as the working solution) in a staining tank for 15 min in a 37 ℃ oven. Observe and find the division phase where chromosomes are evenly separated under a microscope. Capture and analyze the number of chromosomes in 20 metaphases for each iPS cell line (Fig. S2). In total, 10 OSKM + Wdr82 iPSC lines and 5 OSKM iPSC lines were assessed for normal karyotype percentage.
Chimeric mouse experiment
Chimeric mice were produced with a white recipient mouse embryo with an ICR background. Chimeric embryos were obtained via the 8-cell embryo polymerization method and transplanted into a mouse uterus. The chimeric mice were born after 13.5 days in utero via cesarean section upon veterinary anesthesia using avertin (soamyl alcohol and tribromoethanol mixture in an equal proportion, followed by 50 times dilution, and inject at 0.15 times the body weight of the mouse). Because the donor OSKM-Wdr82-iPS cells carried a black C57 background, the adult chimeric mice presented with a black and white coat color (Fig. S3).
RNA-seq
Three sets of biological replicate cell samples collected on reprogramming Day 3, Day 6 and/or Day 9, as well as the original somatic MEF cells, were subjected to RNA-seq. These samples were named as follows: Wdr82-1, Wdr82-2, and Wdr82-3 and Vector-1, Vector-2, and Vector-3 for the cell samples collected at reprogramming Day 3 from the OSKM + Wdr82 and OSKM groups; similarly, Wdr82-4, Wdr82-5, and Wdr82-6 and Vector-4, Vector-5,and Vector-6 were the names of the samples obtained on reprogramming Day 6, and Wdr82-7, Wdr82-8, and Wdr82-9 and Vector-7, Vector-8, and Vector-9 were the names of the samples obtained on Day 9. MEF-1, MEF-2, and MEF-3 were the negative control cells. Genes that were differentially upregulated or downregulated during the reprogramming process were identified based on a cutoff P value < 0.05 and |log2(fold-change)|≥ 1. Heatmaps were drawn using Heatmapper (http://www.heatmapper.ca/). KEGG and Gene Ontology Biological Process analyses were performed using the DAVID gene set enrichment tool. The primer pairs used for qRT‒PCR to validate of the identification of the differentially expressed genes associated with OXPHOS are listed in Table S6.
MMP assay
Mitochondrial membrane potential (MMP) was assessed using the mitochondrial membrane potential assay dye JC-1 according to the manufacturer's protocol (Beyotime, C2006, Nanjing, China). A total of 5 × 105 cells were washed twice with PBS and incubated with JC-1 dye in serum-free medium for 20 min at 37 °C. After washing twice, the cells were analyzed using a BD Accuri C6 flow cytometer or observed under a microscope (IX73, Olympus, Japan). In normal mitochondria, JC-1 aggregates in the mitochondrial matrix to form a polymer, which emits intense red fluorescence (Em = 590 nm), while in cells with lost membrane potential in unhealthy mitochondria, JC-1 is found only as monomers in the cytoplasm, emitting green fluorescence (Em = 529 nm).
ROS detection
ROS generated in cells were detected using a DHE probe (Applygen, C1300-2, Beijing, China). Cells in the supernatant were removed and incubated at 37 °C for 30 min in DHE probe solution diluted with serum-free culture medium. The upper layer of the culture medium was discarded, and the cells were washed with DPBS, and then observed under a microscope (Em = 520 nm).
ATP production quantitation
The ATP amount produced in cells was assessed according to the standard protocol (Beyotime, S0026, Nanjing, China). Cells were lysed in lysis buffer, and then the supernatant was taken for subsequent determination after centrifugation for 5 min at 4 ℃. The supernatant was incubated for 5 min in a proportional ATP detection working solution at room temperature, followed by equal mixing with the standard solution and measurement of the RLU or CPM value with a chemiluminescence meter. The ATP value in OSKM-Wdr82 cells was obtained after normalization of the background and the subsequent OSKM control experiments.
MitoTracker staining assay
Cells on coverslips in a petri dish were removed from the culture medium and then incubated for 20 min in a prewarmed staining solution containing a MitoTracker probe (Thermo Fisher Invitrogen, M22426). The staining solution was then replaced with fresh prewarmed medium, and cells were observed under a fluorescence microscope (Em = 520 nm).
Statistical analyses
All data are reported as the means ± SEMs. The results were analyzed using Student’s t test. Differences were considered statistically significant when P < 0.05. At least three independent experiments were performed in replicates for statistical analyses. *P < 0.05, **P < 0.01 and ***P < 0.001.
Results
Wdr82 increases somatic cell reprogramming efficiency
Since MII oocyte-derived transcription factors might increase the generation of induced pluripotent stem cells [12, 35], three highly expressed factors in MII oocytes that possibly participating in epigenetic modification of Chromatin were selected for analysis, namely, Wdr82 (WD repeat domain 82), Dpy30 (dpy-30 histone methyltransferase complex regulatory subunit), and Taf7 (TATA-box binding protein-associated factor 7), and their fusion expression clones were inserted into a Fuw-TetOn vector backbone. Oct4-GFP/Rosa26-M2rtTA mouse embryonic fibroblasts (OR-MEFs) were used as the sources of somatic cells to induce pluripotent stem cell generation at the basis of the induction by Yamanaka factors (OSKM factors) (Figs. S1, 1A). The effects of these three factors on somatic cell reprogramming were evaluated. The results indicated that among the three factors, Wdr82 overexpression resulted in the most significant promotion of somatic cell reprogramming, and its effects were very stable. Many more Oct4-GFP+ clones were produced during reprogramming induced by OSKM + Wdr82 when compared with that induced with the OSKM control and/or by the addition of Dpy30/Taf7 (Fig. 1B). Moreover, the number of Oct4-GFP+ clones was significantly increased, by 2.1-fold, as compared to that of the OSKM control clones (Fig. 1C and D). These data were confirmed via flow cytometry analysis, which showed that the increase in Oct4-GFP+ clones was 2.9-fold higher than that in the control clones (Fig. 1E). In addition, alkaline phosphatase (AP) staining showed that the number of AP+ clones increased by 3.4-fold (Fig. 1F). These findings indicate that Wdr82 increased somatic cell reprogramming efficiency. Then, the pluripotency of the reprogrammed Oct4-GFP+ clones was further assessed.
Overexpressed Wdr82 increases reprogramming efficiency. A Changes in the morphology of cells during somatic cell reprogramming as observed with bright field microscopy. B The morphology of the clones overexpressing Wdr82, Dpy30, or Taf7 as observed by bright field and fluorescence microscopy. C and D In the reprogramming experiment, overexpressed Wdr82 led to the production of the most GFP+ clones, as shown in the line graph (C) and the histogram (D). E Among the three analyzed factors, by the end of the reprogramming induction period, Wdr82 overexpression produced the highest proportion of Oct4-positive cells, as assessed by flow cytometry. F Among the three factors, Wdr82 overexpression induced the greatest production of AP+ clones at the end of the reprogramming induction period. The number of positive clones is indicated as the mean ± standard error, n = 3; significant difference was determined by t test: *P < 0.05, **P < 0.01
OSKM + Wdr82 reprogrammed cell lines showed pluripotency and chimeric mice were produced
To verify the promoting effect of Wdr82 on mouse somatic cell reprogramming, in this study, Oct4-GFP+ monoclonal cells obtained by reprogramming were selected (denoted as OSKM + Wdr82-iPSCs; Fig. 2A), and then, the pluripotency and differentiation potential of these cells were evaluated. The results showed that the morphology of the OSKM + Wdr82-iPSCs was similar to that of ESCs and that the OSKM + Wdr82-iPSCs expressed Oct4 (a fluorescent reporter gene) (Fig. 2B) and presented a normal karyotype (Fig. S2, Fig. 2C). The OSKM + Wdr82-iPSC line with the correct karyotype was selected for pluripotency assessment. The RNA expression levels of the pluripotent genes Oct4, Sox2, Nanog, Rex1 and Utf1 in the OSKM + Wdr82-iPSC line were comparable to those in ESCs and markedly higher than those in MEFs (Fig. 2D), and the protein expression of the pluripotent markers SOX2, NANOG, and SSEA1 was evident (Fig. 2E). An in vitro embryoid body (EB) differentiation experiment performed via the hanging drop method (Fig. 2F) indicated that the expression of the pluripotency marker genes specific to the germ layers of differentiated EB spheres was markedly enhanced (Fig. 2G), suggesting that the OSKM + Wdr82-iPSC line showed the potential to differentiate into three embryonic germ layers in vitro. A fundamental in vitro chimeric mouse experiment confirmed that the OSKM + Wdr82-iPSC line transmitted genetic material to offspring mouse cells, as indicated by the embryonic genital ridge in the offspring mice expressing the Oct4-GFP gene (Fig. 2H). An assessment of the adult chimeric mice (Figs. 2I and S3) validated that the OSKM + Wdr82-iPSC line showed pluripotency.
Establishment and function verification of the OSKM + Wdr82-iPSC line. A The flow chart indicates the establishment and verification of the OSKM + Wdr82-iPSC line. B OSKM + Wdr82-iPS cell morphology in bright and dark fields. C Karyotype evaluation: The correct karyotype acquisition rate of OSKM + Wdr82-iPSC line was equivalent to that of the OSKM control cells, detailed in Fig. S2; D RT-PCR detection of pluripotency gene (Oct4, Sox2, Nanog, Rex1 and Utf1) expression: The expression levels of these genes in the OSKM + Wdr82-iPSC line were equivalent to that in ESCs (positive controls) and much higher than that in MEFs (negative controls). The fold change in expression is indicated on a log10 scale. E Immunofluorescence staining for detection of pluripotency proteins (SOX2, SSEA1, and NANOG) in the OSKM + Wdr82-iPS cell line. F In vitro EB differentiation of the OSKM + Wdr82-iPSC line. G RT-PCR measurement of the expression of differentiated marker genes. The differentiated OSKM + Wdr82-iPSCs showed higher expression levels of marker genes that were specifically expressed in the three embryonic germ layers, as indicated, and lower expression levels of pluripotency genes than those in the parent iPSCs. The expression fold change is indicated on a log10 scale. H In vivo chimerism experiment: The Oct4-GFP derived from OSKM + Wdr82-iPSC line can be observable in the genital crest, indicating the obtained chimeras mice. I Representative image of adult chimeric mice from two OSKM + Wdr82-iPSC lines, with black and white coat color. * P < 0.05 and ** P < 0.01
In addition, although it promoted somatic cell reprogramming, Wdr82 did not replace any of the four Yamanaka factor genes. To further study the role of Wdr82 in reprogramming induction, a series of induction experiments was performed by replacing each of the OSKM factors with Wdr82, denoted as OSKM, OSK, OSM, OKM and SKM factors, with or without the addition of Wdr82. The results of three biological replicate experiments showed that cells carrying Wdr82 and the four replacement genes expressed Oct4-GFP+ clones in numbers are comparable to the corresponding negative control cells (without Wdr82 overexpression) but in significantly lower numbers than the positive control OSKM cells (Fig. S4A and S4D), suggesting that Wdr82 did not replace the function of the genes encoding of any of the four Yamanaka factors. This result was confirmed by flow cytometric analysis and alkaline phosphatase activity assay carried out at the end of the reprogramming induction period (Fig. S4B and S4C). In summary, Wdr82 cannot be considered a substitute for any of the four Yamanaka factors, although Wdr82 promotes somatic cell reprogramming.
Expression inhibition of Wdr82 significantly suppresses reprogramming efficiency
To confirm the effect of Wdr82 on the reprogramming efficiency, a reprogramming induction experiment in cells which Wdr82 was knocked down via Wdr82-specific short hairpin RNA (shRNA) plasmids was conducted. The results showed that when compared with the OSKM control groups, all three corresponding Wdr82-knockdown constructs significantly inhibited reprogramming efficiency (Figs. S5A, 3A) in three replicate experiments. Moreover, the number of Oct4-GFP+ clones after Wdr82 knockdown was markedly reduced through the reprogramming period, with the inhibitory reprogramming efficiency induced by shWdr82-4 being particularly notable (Figs. 3B and C, S5B). These findings were confirmed via both flow cytometry and AP staining analysis (Figs. 3D, S5C and S5D). In general, knocking down Wdr82 significantly inhibited somatic cell reprogramming efficiency.
Knocking down Wdr82 inhibits somatic cell reprogramming. A The morphology of OSKM + shWdr82 (knockdown) reprogrammed cells and the OSKM control cells. B When compared with the number among OSKM control cells, knocking down Wdr82 (OSKM + shWdr82) significantly reduced the number of Oct4-positive clones. C The proportion of Oct4 + cells after Wdr82 was knocked down was significantly reduced compared with that in the OSKM control cell group. D When compared with that of the OSKM control cells, the AP+ clone number was significantly reduced. * P < 0.05 and ** P < 0.01
The promoting effect of Wdr82 on reprogramming efficiency was mediated by inhibiting OXPHOS activity, suggesting a regulatory role in cellular energy metabolism.
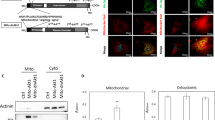
To explore the biological pathways by which Wdr82 increases reprogramming efficiency, a high-throughput transcriptome sequencing (RNA-seq) experiment was performed during the reprogramming process on Days 3, 6 and 9, with corresponding negative control cells (no Wdr82 overexpression) and source cell MEFs included. A heatmap of the Pearson correlation coefficients indicated that the three MEF samples showed completely different RNA expression patterns and that the three biological replicates of each reprogramming sample showed reliable and closely associated variable clusters (Fig. 4A). The results of a principal component analysis (PCA) suggested that the gene expression patterns in reprogrammed cells gradually changed to follow a sloping curve and that, compared to the respective control cells (OSKM + Vector-N), the reprogrammed cells with Wdr82 (OSKM + Wdr82-N) were more concentrated in clusters (solid-lined ovals) (Fig. 4B). The differentially expressed genes (up- and downregulated) on reprogramming Day 9 were much more abundant than those on Day 6 and/or Day 3 (Fig. S6A), which was largely consistent with the morphological observations made during cell reprogramming for approximately 9 days, which showed an increase in colony numbers and green fluorescence. To explore the gene function and biological pathways of Wdr82 in reprogramming, a joint GO and KEGG analysis was conducted based on the differentially expressed genes identified on reprogramming Day 9, and the results indicated that these genes were mainly enriched in oxidative phosphorylation (OXPHOS) and ribosomes (Fig. S6B). Moreover, two biological processes were enriched as determined via an analysis of the protein−protein interaction network (Fig. S6C). The expression of genes involved in oxidative phosphorylation was validated by quantitative RT-PCR measurements; these genes included Ndufas, Ndufbs, Coxs and/or Atpases, were significantly suppressed on reprogramming Day 9 when compared with their expression in the OSKM control cells without Wdr82 addition (Fig. S6D).
Transcriptome analysis of OSKM + Wdr82 cells during reprogramming induction suggested that Wdr82 promotes somatic cell reprogramming by inhibiting OXPHOS. A Heatmap showing the correlation coefficients among samples; Vector-1, Vector-2, Vector-3, ad Wdr82-1, Wdr82-2, and Wdr82-3 represent cell samples collected from the OSKM and OSKM + Wdr82 groups, respectively, on reprogramming Day 3. Similarly, Vector-4, Vector-5, Vector-6, Wdr82-4, Wdr82-5, and Wdr82-6 samples were obtained on reprogramming Day 6, and Vector-7, Vector-8, Vector-9, Wdr82-7, Wdr82-8, and Wdr82-9 samples were collected on Day 9. MEF-1, MEF-2, and MEF-3 were used as the negative controls. B Principal component analysis results showing that the expression pattern in the MEFs was markedly different from that of the cells reprogrammed for 3, 6 and/or 9 days. In the reprogrammed cells, the expression patterns in both the OSKM and OSKM + Wdr82 groups gradually changed throughout the reprogramming process, following an upward sloping curve. Samples in the enclosed solid and dotted line represent samples from OSKM + Wdr82 (Wdr82) and OSKM (Ctrl) cells, respectively. C The expression of genes involved in glycolysis (Hk2, Pk2, and Pfk) and OXPHOS (Ndufc1, Ndufb7, Atp5e, Cpt1α, and Cpt2) in addition to that of Wdr82 was inhibited during reprogramming (Day 6 ~ Day 9), while fermentation gene expression associated with induced lactate production (Ldhα) was significantly enhanced as compared to that in the OSKM-only control group. D A MitoTracker assay indicated that the number of mitochondria in cells after the addition of Wdr82 slowly decreased compared to that in the OSKM control group during cell reprogramming (Day 3 ~ Day 9). Red fluorescence represents mitochondria in the cytoplasm. E Quantitation of the MitoTracker assay data. F ROS production in reprogrammed cells on Day 9 was decreased after the addition of Wdr82, and the quantification results were shown as in the column chart. * P < 0.05, ** P < 0.01 and *** P < 0.001
This finding suggested that in the cell reprogramming process, the energy metabolism pathway was specifically reprogrammed, as indicated by the significant inhibition of OXPHOS and aerobic metabolism in the reprogrammed cells. Therefore, a series of experiments to assess the energy metabolism pathway during somatic reprogramming was carried out. First, in addition to that of the genes involved in OXPHOS (Ndufc1, Ndufb7, Atp5e, Cpt1α, and Cpt2), the expression of genes involved in glycolysis (Hk2, Pk2, and Pfk) was inhibited during reprogramming (Day 6–Day 9, Fig. 4C). In contrast, the expression of the gene involved in fermentation-inducing lactate production (Ldhα) was significantly enhanced. Second, a MitoTracker assay indicated that the number of mitochondria within the cell after the addition of Wdr82 was slowly decreased as compared to that in the OSKM control cells during cell reprogramming (Day 3–Day 9, Fig. 4D and E). Third, a JC-1 mitochondrial membrane potential assay indicated that cells treated with Wdr82 + OSKM showed significantly more unhealthy/damaged mitochondria than those treated with OSKM only during the reprogramming period, especially on Day 9 (Figure S6E and S6F). Moreover, gradually decreased ROS accumulation was accompanied by slowly inhibited mitochondrial function (Fig. 4F) in the cells undergoing reprogramming (Day 9). All of the aforementioned evidence suggested that Wdr82 promotes somatic cell reprogramming by inhibiting OXPHOS in mitochondria but enhancing fermentation (Fig. 5). The study provides a link between mitochondrion metabolism and cell reprogramming efficiency.
Abundantly expressed factor Wdr82 in oocytes can facilitate iPSCs generation by inhibiting mitochondrial oxidative phosphorylation. Wdr82, a C-terminal domain-binding protein that recruits the Setd1A/B histone H3K4 methyltransferase complex and phosphorylates RNA polymerase II, suppresses the transcrptions of genes (Nudf etc.) involved in OXPHOS in mitochondria in the early stage of the reprogramming, thereby inhibiting OXPHOS and transferring the basis of the carbon energy source from FAO to glycolysis (lactate), promoting fermentation, and ultimately mediating cell fate reprogramming. The process is accompanied with changes of mitochondrial quantity and structure. The study provides evidence showing that metabolic switching mediated through metabolic function, epigenetic modifications and/or gene expression is critical for cell fate determination
Discussion
In this study, three highly expressed factors in MII phase oocytes, Wdr82, Dpy30 and Taf7, were selected as possible candidates for promoting somatic cell reprogramming. Our data showed higher reprogramming efficiency after the addition of Wdr82 than that of the four Yamanaka factors only (Fig. 1), providing scientific evidence for the hypothesis that abundantly expressed oocyte factors promote the generation of induced pluripotent stem cells and/or embryonic development [12, 35]. In particular, iPSC quality after the addition of Wdr82 was shown to be excellent, with pluripotency and heritability to chimeric offspring mice (Fig. 2).
In terms of the molecular mechanisms underlying how oocyte factors regulate the reprogramming process may differ depending on the factor. Our investigation revealed that Wdr82 promoted programming efficiency via OXPHOS inhibition at the early stage of programming (Fig. 4). Meanwhile, the addition of Wdr82 did enhanced the H3K4me3 level in reprogrammed cells at day 6 and day 9 (Fig. S7), confirming its recruitment role in the Setd1A/B complex, a H3K4me3 methyltransferase complex, during the reprogramming process. Moreover, Wdr82 failed to be a suitable replacement for any of the four Yamanaka factors (Fig. S4), suggesting its unique roles in the regulation of cell fate determination. With regard to OXPHOS during reprogramming, increasing evidence has suggested that a cell fate transition is usually accompanied by reconstruction of the energy metabolism pathway. The metabolism of somatic cells and stem cells is very different. As the main sites of cell respiration and energy production, mitochondria play important roles in cellular energy metabolism. Somatic cells heavily rely on mitochondrial oxidative phosphorylation as the main source of energy [9], while ESCs rely on glycolysis for ATP production-based energy [7]. The upregulation of glycolytic enzymes and the downregulation of electron transport chain subunits switch mitochondrial oxidative metabolism into a glycolysis-dependent state, which is the basis for acquiring a pluripotent phenotype (Fig. 5) [11, 34, 40]. For example, the histone acetyltransferase MOF directly activates the fatty acid oxidation (FAO) pathway in mitochondria, thereby blocking quiescence in ESCs, which show ground state pluripotency [20]. Similarly, a protein on the surface of lipid droplets (LDs), perilipin 2 (Plin2), accelerates ESC decline in pluripotency by enhancing lipidomic remodeling and histone acetylation [39], indicating a mechanism linking LD homeostasis to mitochondrial remodeling (the development of mitochondrial cristae and FAO) and epigenetic regulation [26,27,28]. FAO provides the carbon source for fueling mitochondrial respiration, that is, OXPHOS. In summary, the importance of energy metabolism is not only essential for cell survival and proliferation, but is also increasingly recognized in pluripotency maintenance and cell fate determination [26, 29]. Here, we found that Wdr82 directly modifies the expression of genes involved in mitochondrial OXPHOS, thereby transferring the basis of the carbon energy source from FAO to glycolysis (lactate), inhibiting OXPHOS, promoting fermentation (Fig. 4), and ultimately mediating cell fate reprogramming (Fig. 5).
Moreover, Wdr82, a scaffold protein, is a component of the Setd1A/B complex, a H3K4me3 methyltransferase complex related to gene transcription activation, that regulates the expression of many genes in early embryonic development. Studies have reported that knocking down Wdr82 in mESCs significantly reduced the level of H3K4me3, slows the cell proliferation rate and inhibits cell cycle progression through the p53−p21 pathway [3]. The loss of Wdr82 specificity leads to a significant decrease in the rate of H3K4 trimethylation and the level of actively transcribed genes in chromatin [38]. Oct4 is a downstream factor of the Setd1A/B complex, and the absence of Wdr82 reduces the Setd1A/B complex level, which hinders the growth and development of an embryo, in some cases leading to embryonic death [3]. Wdr82 is also present in the protein phosphatase 1 (PP1) complex. The PP1 nuclear targeting subunit (Pnuts) is necessary for PP1 recruitment to active transcription sites, and it dephosphorylates the carboxy-terminal repeat domain of RNA polymerase II (Pol II) at active transcription sites [1]. Wdr82 inhibits transcription−replication conflict by promoting the degradation of Pol II on chromatin and reducing its residence time. Pnut-PP1-mediated dephosphorylation of the carboxy-terminal domain of Pol II led to results similar to Wdr82 [22]. In summary, Wdr82 activates the transcription of abundant downstream genes, which may be associated with metabolic functions, epigenetic modification and/or gene repression, by recruiting transcription factors that bind to Pol II.
Knocking down Wdr82 significantly decreased the H3K4 trimethylation rate in mESCs, causing considerable delays in the G1 phase and a slowed proliferation rate. The expression of the cell cycle-dependent kinase inhibitor p21 and tumor suppressor gene p53 was significantly upregulated, indicating that inhibition of the cell cycle by Wdr82 knockdown was realized through the p53−p21 pathway. Dai et al. reported that Akt directly phosphorylates Oct4 to regulate the formation of Oct4/Sox2 heterodimers and promotes p300-mediated acetylation of Oct4, Sox2 and Klf4, thereby indirectly promoting iPSC formation [8]. P53 can directly control cell death and mitochondrial respiration and can act on the Mdm2, Mdm4, Arf and Bmi1 factors in mitochondria. Mdm4 is related to the control of cell death. In summary, the p53 pathway is closely related to mitochondrial function and fine tunes metabolic processes [21]. Akt phosphorylates Mdm2 and promotes the degradation of p53 by the proteasome [32], and the caspase cascade induced by p53 accelerates the degradation of Mdm2 and Akt [13]. Therefore, Wdr82 is thought to exert its regulatory effect via the Pi3k−Akt pathway, thereby inhibiting the p53 signaling pathway. P21, a target gene downstream of p53, inhibits the expression of genes associated with mitochondrial oxidative phosphorylation and possibly that of epigenetic modifications and/or pluripotent genes.
Recent evidence supports the idea that mitochondria are signaling organelles that dictate stem cell fate and function. Mitochondrial metabolism is linked to the TCA cycle/ATP production and aerobic metabolism to support cell fate determination through a wide variety in regulatory cellular functions [4]. Moreover, cell fate decisions are susceptible to intrinsic metabolic bias imposed by selectively inherited mitochondria [10, 26]. Moreover, lactate, as a byproduct of glycolysis, which is upregulated in response to hypoxia (anaerobic metabolism), has been identified by researchers of the Warburg effect and has recently been considered the primary substrate of lactylation, an important posttranslational modification, thereby playing a critical role in regulating homeostatic and pathological processes [26,27,28, 41]. Therapeutics based on mitochondrion metabolism remodeling may be attractive strategies to improve stem cell function to attenuate disease and/or promote healthy aging due to their fate-determining chromatin epigenetic modifications, hypoxic transcriptional responses and/or immune functions [2, 30, 42].
Conclusion
Overall, a transcription factor abundantly expressed in MII oocytes, Wdr82, can increase the efficiency of somatic cell reprogramming induction. A Wdr82-expressing iPSC line showed pluripotency with the potential to differentiate into three germ layers and to transmit genetic material to chimeric offspring. The significant suppression of OXPHOS in mitochondria underlies the molecular mechanism by which Wdr82 promotes the efficiency of somatic cell reprogramming. Our study provides a link between mitochondrion metabolism and cell fate determination.
Availability of data and materials
The RNA-seq data used to support the findings of this study are deposited in GEO/NCBI and publicly available with ID of PRJNA975608.
Abbreviations
- AP:
-
Alkaline phosphatase
- ATP:
-
Adenosine triphosphate
- Atp5e:
-
ATP synthase H + transporting mitochondrial F1 complex epsilon subunit
- c-Myc:
-
MYC proto-oncogene, a bHLH transcription factor
- Cpt1α:
-
Carnitine O-palmitoyltransferase 1
- Cpt2:
-
Carnitine O-palmitoyltransferase 2
- Dpy30:
-
Dpy-30 histone methyltransferase complex regulatory subunit
- FAO:
-
Fatty acid oxidation
- Glis1:
-
GLIS family zinc finger 1
- H1foo:
-
Oocyte-specific H1 histone
- Hk:
-
Hexokinase
- iPSC:
-
Induced pluripotent stem cell
- Klf4:
-
Kruppel like factor 4
- Mdm2/4:
-
MDM2/4 proto-oncogene
- MEF:
-
Mouse embryonic fibroblast
- MMP:
-
Mitochondrial membrane potential
- Nanog:
-
Nanog homeobox
- Nduf:
-
NADH dehydrogenase (ubiquinone) 1 subunit, mitochondrial
- Obox1:
-
Oocyte-specific homeobox 1
- Oct4:
-
POU domain, class 5, transcription factor 1
- OSKM:
-
Oct4, Sox2, Klf4 and c-Myc
- OXPHOS:
-
Oxidative phosphorylation
- Pf:
-
Pyruvate kinase
- Pfk:
-
ATP-dependent 6-phosphofructokinase
- Rex1:
-
RNA exonuclease 1
- ROS:
-
Reactive oxygen species
- Setd1b:
-
SET domain-containing 1B, histone lysine methyltransferase
- Sox2:
-
SRY-box transcription factor 2
- Taf7:
-
TATA-box-binding protein associated factor 7
- TCA cycle:
-
Tricarboxylic acid cycle
- Tcl1:
-
T-cell lymphoma breakpoint 1
- Tet1:
-
Tet methylcytosine dioxygenase 1
- Utf1:
-
Undifferentiated embryonic cell transcription factor 1
- Wdr5:
-
WD repeat domain 5
- Wdr82:
-
WD repeat domain 82
- Yamanaka factors:
-
Oct4, Sox2, Klf4 and c-Myc
- Zscan4:
-
Zinc finger and SCAN domain containing 4
References
Austenaa LMI, Barozzi I, Simonatto M, Masella S, Chiara GD, Ghisletti S, Curina A, Wit ED, Bouwman BAM, Pretis SD, Piccolo V, Termanini A, Prosperini E, Pelizzola M, Laat WD, Natoli G (2015) Transcription of mammalian cis-regulatory elements is restrained by actively enforced early termination. Mol Cell 60(3):460–474
Bargiela D, Burr SP, Chinnery PF (2018) Mitochondria and hypoxia: metabolic crosstalk in cell-fate decisions. Trends Endocrinol Metab 29(4):249–259
Bi Y, Lv Z, Wang Y, Hai T, Huo R, Zhou ZM, Zhou Q, Sha JH (2011) WDR82, a key epigenetics-related factor, plays a crucial role in normal early embryonic development in mice. Biol Reprod 84(4):756–764
Chakrabarty RP, Chandel NS (2021) Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell 28(3):394–408
Chen JY, Gao YW, Huang H, Xu K, Chen X, Jiang YH, Li H, Gao S, Tao Y, Wang H, Zhang Y, Wang HL, Cai T, Gao SR (2015) The combination of Tet1 with Oct4 generates high-quality mouse-induced pluripotent stem cells. Stem Cells 33(3):686–698
Cheng ZL, Zhang ML, Lin HP, Gao C, Song JB, Zheng ZH, Li LP, Zhang YN, Shen XQ, Zhang H, Huang ZH, Zhan WQ, Zhang C, Hu X, Sun YP, Jiang LB, Sun L, Xu YH, Yang C, Ge YL, Zhao Y, Liu XG, Yang H, Liu PY, Guo X, Guan KL, Xiong Y, Zhang ML, Ye D (2020) The Zscan4-Tet2 transcription nexus regulates metabolic rewiring and enhances proteostasis to promote reprogramming. Cell Rep 32(2):107877
Chung S, Dzeja PP, Faustino RS, Perez-Terzic C, Behfar A, Terzic A (2007) Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med Suppl 1:S60–S67
Dai XP, Liu PD, Lau AW, Liu YY, Inuzuka H (2014) Acetylation-dependent regulation of essential iPS-inducing factors: a regulatory crossroad for pluripotency and tumorigenesis. Cancer Med 3(5):1211–1224
Deberardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB (2008) The biology of Cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7(1):11–20
Dohla J, Kuuluvainen E, Gebert N, Amaral A, Englund JI, Gopalakrishnan S, Konovalova S, Nieminen AI, Salminen ES, Muñumer RT, Ahlqvist K, Yang Y, Bui H, Otonkoski T, Käkelä R, Hietakangas V, Tyynismaa H, Ori A, Katajisto P (2022) Metabolic determination of cell fate through selective inheritance of mitochondria. Nat Cell Biol 24(2):148–154
Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikaeda Y, Perez-Terzic C, Terzic A (2011) Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metab 14(2):264–271
Gao YW, Liu XY, Tang B, Li C, Kou ZH, Li L, Liu WQ, Wu Y, Kou XC, Li JY, Zhao YH, Yin JQ, Wang H, Chen S, Liao LJ, Gao SR (2017) Protein expression landscape of mouse embryos during pre-implantation development. Cell Rep 21(13):3957–3969
Gottlieb TM, Rony Seger JFML, Taya Y, Oren M (2002) Cross-talk between Akt, p53 and Mdm2: possible implications for the regulation of apoptosis. Oncogene 21(8):1299–1303
Guo SQ, Zi XY, Schulz VP, Cheng JJ, Zhong M, Koochaki SH, Megyola CM, Pan XH, Heydari K, Weissman SM, Gallagher PG, Krause DS, Fan R, Lu J (2014) Nonstochastic reprogramming from a privileged somatic cell state. Cell 156(4):649–662
Hernandez C, Wang Z, Ramazanov B, Tang Y, Mehta S, Dambrot C, Lee YW, Tessema K, Kumar I, Astudillo M, Neubert TA, Guo SQ, Ivanova NB (2018) Dppa2/4 facilitate epigenetic remodeling during reprogramming to pluripotency. Cell Stem Cell 23(3):396–411
Jiang JJ, Lv WJ, Ye XY, Wang LB, Zhang M, Yang H, Okuka M, Zhou CK, Zhang X, Liu L, Li JS (2013) Zscan4 promotes genomic stability during reprogramming and dramatically improves the quality of iPS cells as demonstrated by tetraploid complementation. Cell Res 23(1):92–106
Jullien J, Miyamoto K, Pasque V, Allen GE, Bradshaw CR, Garrett NJ, Halley-Stott RP, Kimura H, Ohsumi K, Gurdon JB (2014) Hierarchical molecular events driven by oocyte-specific factors lead to rapid and extensive reprogramming. Mol Cell 55(4):524–536
Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, Wahl GM, Izpisúa Belmonte JC (2009) Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 460(7259):1140–1144
Khaw SL, Min-Wen C, Koh CG, Lim B, Shyh-Chang N (2015) Oocyte factors suppress mitochondrial polynucleotide phosphorylase to remodel the metabolome and enhance reprogramming. Cell Rep 12(7):1080–1088
Khoa LTP, Tsan YC, Mao FB, Kremer DM, Sajjakulnukit P, Zhang L, Zhou B, Tong X, Bhanu NV, Choudhary C, Garcia BA, Yin L, Smith GD, Saunders TL, Bielas SL, Lyssiotis CA, Dou YL (2020) Histone acetyltransferase MOF blocks acquisition of quiescence in ground-state escs through activating fatty acid oxidation. Cell Stem Cell 27:1–18
Lahalle A, Lacroix M, Blasio CD, Cisse MY, Linares LK, Cam LL (2021) The p53 pathway and metabolism: the tree that hides the forest. Cancers 13(1):133
Landsverk HB, Sandquist LE, Bay LTE, Steurer B, Campsteijin C, Landsverk OJB, Marteijin JA, Petermann E, Trinkle-Mulcahy L, Syljuasen RG (2020) WDR82/PNUTS-PP1 prevents transcription-replication conflicts by promoting rna polymerase ii degradation on chromatin. Cell Rep 33(9):108469
Le R, Huang Y, Zhang Y, Wang H, Lin J, Dong Y, Li Z, Guo M, Kou X, Zhao Y, Chen M, Zhu Q, Zhao A, Yin J, Sun J, Su Z, Shi K, Gao Y, Chen J, Liu W, Kang L, Wang Y, Li C, Liu X, Gao R, Wang H, Ju Z, Gao S (2021) Dcaf11 activates Zscan4-mediated alternative telomere lengthening in early embryos and embryonic stem cells. Cell Stem Cell 28(4):732-747.e9
Le RR, Kou ZH, Jiang YH, Li M, Huang B, Liu WQ, Li H, Kou XC, He WZ, Rudolph KL, Ju ZY, Gao SR (2014) Enhanced telomere rejuvenation in pluripotent cells reprogrammed via nuclear transfer relative to induced pluripotent stem cells. Cell Stem Cell 14(1):27–39
Lee JH, Skalnik DG (2008) Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A Histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol 28(2):609–618
Li L, Chen K, Wang T, Wu Y, Xing G, Chen M, Hao Z, Zhang C, Zhang J, Ma B, Liu Z, Yuan H, Liu Z, Long Q, Zhou Y, Qi J, Zhao D, Gao M, Pei D, Nie J, Ye D, Pan G, Liu X (2020) Glis1 facilitates induction of pluripotency via an epigenome-metabolome-epigenome signalling cascade. Nat Metab 2(9):882–892
Li W, Long Q, Wu H, Zhou Y, Duan L, Yuan H, Ding Y, Huang Y, Wu Y, Huang J, Liu D, Chen B, Zhang J, Qi J, Du S, Li L, Liu Y, Ruan Z, Liu Z, Liu Z, Zhao Y, Lu J, Wang J, Chan WY, Liu X (2022) Nuclear localization of mitochondrial TCA cycle enzymes modulates pluripotency via histone acetylation. Nat Commun 13(1):7414
Li XL, Yang YY, Zhang B, Lin XT, Fu XX, An Y, Zou YL, Wang JX, Wang ZB, Yu T (2022) Lactate metabolism in human health and disease. Signal Transduct Target Ther 7(1):305
Liu WW, Chen GK (2021) Regulation of energy metabolism in human pluripotent stem cells. Cell Mol Life Sci 78:8097–8108
Martinex-Reyes I, Chandel NS (2020) Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 11(1):102
Penalosa-Ruiz G, Bousgouni V, Gerlach JP, Waarlo S, Ven JV, Veenstra TE, Silva JCR, Heeringen SJV, Bakal C, Mulder KW, Veenstra GJC (2019) WDR5, BRCA1, and BARD1 co-regulate the dna damage response and modulate the mesenchymal-to-epithelial transition during early reprogramming. Stem Cell Rep 12(4):743–756
Schaefer T, Steiner R, Lengerke C (2020) SOX2 and p53 expression control converges in PI3K/AKT signaling with versatile implications for stemness and cancer. Int J Mol Sci 21(14):4902
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126(4):663–676
Wanet A, Arnould T, Najimi M, Renard P (2015) Connecting mitochondria, metabolism, and stem cell fate. Stem Cells Dev 24(17):1957–1971
Wang SF, Kou ZH, Jing ZY, Guo XZ, Dong MQ, Wilmut I, Gao SR (2010) Proteome of mouse oocytes at different developmental stages. Proc Natl Acad Sci USA 107(41):17639–17644
Wei TY, Chen W, Wang XK, Zhang M, Chen JY, Zhu SC, Chen L, Yang DD, Wang GY, Jia WW, Yu YY, Dun T, Wu MJ, Liu HQ, Gao SR, Kang JH (2015) An HDAC2-TET1 switch at distinct chromatin regions significantly promotes the maturation of pre-iPS to iPS cells. Nucl Acids Res 43(11):5409–5422
Wu L, Wu Y, Peng B, Hou ZZ, Dong Y, Chen K, Guo MY, Li H, Chen X, Kou XC, Zhao YH, Bi Y, Wang YX, Wang H, Le RR, Kang L, Gao SR (2017) Oocyte-specific homeobox 1, Obox1, facilitates reprogramming by promoting mesenchymal-to-epithelial transition and mitigating cell hyperproliferation. Stem Cell Rep 9(5):1692–1705
Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A (2008) Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 28(24):7337–7344
Wu Y, Chen KS, Li LP, Hao ZH, Wang TY, Liu Y, Xing GS, Liu ZC, Li HY, Yuan H, Lu JH, Zhang C, Zhang JY, Zhao DY, Wang JW, Nie JF, Ye D, Pan GJ, Chan WY, Liu XG (2022) Plin2-mediated lipid droplet mobilization accelerates exit from pluripotency by lipidomic remodeling and histone acetylation. Cell Death Differ 29(11):2316–2331
Xu X, Duan S, Yi F, Ocampo A, Liu GH, Izpisua Belmonte JC (2013) Mitochondrial regulation in pluripotent stem cells. Cell Metab 18(3):325–332
Zhang D, Tang ZY, Huang H, Zhou GL, Cui C, Weng YJ, Liu WC, Kim S, Lee S, Perez-Neut M, Ding J, Czyz D, Hu R, Ye Z, He MM, Zheng YG, Shuman HA, Dai LZ, Ren B, Roeder RG, Becker L, Zhao YM (2019) Metabolic regulation of gene expression by histone lactylation. Nature 574(7779):575–580
Zhang HB, Menzies KJ, Auwerx J (2018) The role of mitochondria in stem cell fate and aging. Development 145(8):dev143420
Funding
The work was supported by the National Key R&D Program of China (2021YFD1200903 and 2021YFD1200404), the Key Project of Agricultural Fine Breeding of Shandong Province (2019LZGC011 and 2016LZGC030), the Natural Science Foundation of Shandong Province (ZR2020MC166 and ZR2013CM013) and the National Natural Science Foundation of China (31721003). The funding body did not play a role in the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Author information
Authors and Affiliations
Contributions
G.C., J.Z., K.S. and S.G. conceived the project; J.Z., G.C., J.S., X.K., Z.S., Y.X., T.L., L.S., W.L., and X.W. performed all the experiments. J.Z., G.C., T.L., L.S., W.L. and N.W. analyzed the data. K.S., S.G., G.C., J.Z. and X.W. wrote the paper. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest concerning this article.
Ethical approval and consent to participate
Animal maintenance and experimental procedures were performed according to the ethical approval information.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cui, G., Zhou, J., Sun, J. et al. WD repeat domain 82 (Wdr82) facilitates mouse iPSCs generation by interfering mitochondrial oxidative phosphorylation and glycolysis. Cell. Mol. Life Sci. 80, 218 (2023). https://doi.org/10.1007/s00018-023-04871-z
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-023-04871-z