Abstract
In INF2—a formin linked to inherited renal and neurological disease in humans—the DID is preceded by a short N-terminal extension of unknown structure and function. INF2 activation is achieved by Ca2+-dependent association of calmodulin (CaM). Here, we show that the N-terminal extension of INF2 is organized into two α-helices, the first of which is necessary to maintain the perinuclear F-actin ring and normal cytosolic F-actin content. Biochemical assays indicated that this helix interacts directly with CaM and contains the sole CaM-binding site (CaMBS) detected in INF2. The residues W11, L14 and L18 of INF2, arranged as a 1-4-8 motif, were identified as the most important residues for the binding, W11 being the most critical of the three. This motif is conserved in vertebrate INF2 and in the human population. NMR and biochemical analyses revealed that CaM interacts directly through its C-terminal lobe with the INF2 CaMBS. Unlike control cells, INF2 KO cells lacked the perinuclear F-actin ring, had little cytosolic F-actin content, did not respond to increased Ca2+ concentrations by making more F-actin, and maintained the transcriptional cofactor MRTF predominantly in the cytoplasm. Whereas expression of intact INF2 restored all these defects, INF2 with inactivated CaMBS did not. Our study reveals the structure of the N-terminal extension, its interaction with Ca2+/CaM, and its function in INF2 activation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Formins are a widespread family of proteins involved in polymerizing monomeric globular actin into linear filamentous actin (F-actin) [1, 2]. Formins possess two characteristic domains: a formin homology (FH) 2 domain, which catalyzes actin polymerization, and an FH1 domain, which binds profilin to provide G-actin to the FH2 domain to form F-actin. Most formins contain a diaphanous inhibitory domain (DID) at the N-terminal region that interacts with a diaphanous autoregulatory domain (DAD) located at the C-terminal region to close the molecule in an inactive state [3]. In the case of the formin mDia1, there is an N-terminal extension to the DID with a domain, called the G domain, that forms a bipartite surface in conjunction with the DID to constitute the GTPase-binding domain (GBD), which is responsible for binding the GTPase Rho in its active GTP-loaded form [4,5,6]. The binding of Rho to the GBD disrupts the DID–DAD interaction and opens mDia1 in a catalytically active form [4, 5, 7]. However, although other formins also bind Rho-family GTPases, this mechanism of activation is not shared by all DID and DAD-containing formins [8].
INF2 is a DID and DAD-containing formin linked to inherited human disease [9]. Pathogenic INF2 mutations are autosomal dominant and produce focal segmental glomerulosclerosis [10, 11], which causes a partial loss of podocytes that may end in renal failure. Depending on the specific mutation, focal segmental glomerulosclerosis is combined or not with Charcot–Marie–Tooth disease [12], which is a neurological disorder affecting the functioning of the peripheral nerves leading to progressive distal muscle weakness [13]. The overall domain organization of INF2 is similar to that of mDia1, except that it has an N-terminal extension of unknown structure and function that is shorter than, and has no homology with, that of mDia1 [14, 15]. Elevation of intracellular Ca2+ levels triggers a rapid remodeling of the actin cytoskeleton in an INF2 expression-dependent manner [16, 17]. This remodeling is mediated by the association of calmodulin (CaM) with INF2 [17, 18], but it was not established whether or not the two proteins interact directly. In this work we have addressed the structure and function of the N-terminal extension of INF2 and found it to be organized into two α-helices, the first of which interacts directly with INF2 and contains the sole CaM-binding site (CaMBS) of the INF2 molecule. Inactivation of this site by the mutation of critical residues abolished the actin polymerization response of INF2 to augmented Ca2+ levels. In conclusion, whereas mDia1 is activated by the binding of Rho to the bipartite CaM-binding site at the N-terminal region, the direct binding of Ca2+/CaM to the CaMBS in the first α-helix of the N-terminal extension does so for the INF2 molecule.
Results and discussion
The N-terminal extension of INF2 is organized into two α-helices
Human INF2 has a 35-amino-acid-long extension at the N-terminal end whose structure and function are unknown, while the extension present in mDia1 is longer and contains a part, called the G domain, of the Rho GTPase-binding site (Fig. 1A). The NMR study of a synthetic peptide comprising the full N-terminal extension of INF2 (INF2 2–36) revealed two α-helices, respectively, spanning amino acids 3–17 and the second spanning amino acids 25–31, which were separated by a short unstructured region (Figs. 1B and S1A–D). The two helices were well defined (Fig. S1C, D and Table S1), but not their relative orientation with respect to one another (Fig. S1B). A synthetic peptide comprising only residues 2-19 was shown to form a α-helical structure identical to the first helix of the 2–36 peptide (Fig. S1E, F).
The N-terminal extension of INF2 contains two α-helices. A INF2 has an extension N-terminal to the DID that is shorter than that of mDia1 and lacks a G domain. G (also known as GBDN) domain necessary for Rho binding that is adjacent to the N terminus of the DID, GBD GTPase-binding site, DAD diaphanous autoinhibitory domain, DID diaphanous inhibitory domain, FH1 and FH2 formin homology 1 and 2, respectively. B Ribbon representation of the NMR solution structure of the N-terminal extension of INF2. Backbone atoms are shown in black. K and R side chains are displayed in blue, D and E side chains in red, and all others in green
The first α-helix of the N-terminal extension of INF2 is required for actin homeostasis
For functional studies, we used Madin–Darby canine kidney (MDCK) cells, which are a paradigm of epithelial cells [19]. To identify alterations in the actin cytoskeleton from the lack of INF2 expression, we compared INF2 KO cells [20] expressing the indicated proteins (Fig. 2A). INF2 KO cells lacked the thin ring of F-actin that surrounds the nucleus of MDCK cells under our cell culture conditions [21, 22] and had a lower cytosolic F-actin content than did the control cells (Fig. S2A–C). Consistent with previous findings in HeLa KO cells [17], expression of either of the two isoforms of INF2, INF2-1 and INF2-2 (also known as INF2 CAAX and INF2 nonCAAX, respectively), restored these defects (Fig. 2B–D). However, INF2-1 was more efficient than INF2-2 in polymerizing actin at the perinuclear ring, probably because INF2-1 is associated with the endoplasmic reticulum whereas INF2-2 is cytoplasmic [23]. To analyze the function of the N-terminal extension, we chose INF2-1, hereafter called INF2 for simplicity, to express deletion mutants lacking both helices (INF2 Δ2-30) or only the first of them (INF2 Δ2–18). It is of note that neither of the deletion mutants rescued the normal F-actin phenotype (Fig. 2B–D). The levels of expression in INF2 KO cells of the exogenous INF2 proteins assayed were 6–9 times higher than those of endogenous INF2 in control cells (Fig. S2D, E). No significant differences were found in the expression levels between the exogenous INF2 proteins (Fig. S2D, F). In conclusion, the first α-helix of the N-terminal extension of INF2 is required to ensure the normal distribution and cellular content of F-actin.
The first of α-helix of INF2 is required for the normal distribution and content of F-actin. A Schematic of the INF2 proteins expressed. B–D INF2 KO cells or INF2 KO cells expressing mCherry alone or mCherry fusions of the indicated INF2 proteins were stained for F-actin. Nuclei were visualized with DAPI. An enlargement of the DAPI and F-actin staining corresponding to the boxed regions is shown in the rightmost panels. Scale bar, 10 μm (B). Box plots showing the intensity of F-actin staining at the perinuclear region (C) and the cytosol (D) relative to that of control INF2 KO cells (> 140 cells were analyzed for each experimental condition; three independent experiments; ns not significant; ***p < 0.001)
The first α-helix of the N terminus of INF2 binds Ca2+/calmodulin
Elevation of intracellular Ca2+ levels triggers a rapid and transient remodeling of F-actin in an INF2 expression-dependent manner in 3T3 fibroblasts [16] and MDCK cells [17]. Since INF2 binds to CaM in the presence, but not in the absence, of Ca2+ [17, 18, 24], CaM appears to mediate this response. To investigate whether the actin alterations observed in INF2 KO cells expressing INF2 proteins with N-terminal deletions are correlated with the loss of binding of CaM, we carried out pull-down assays with fragments of the N-terminal region of INF2 fused to GST to analyze GFP-CaM binding (Fig. 3A). We observed Ca2+-dependent binding to the INF2 fragment encompassing amino acids 2–340, which contains the DID, the dimerization domain, and the entire N-terminal extension (Fig. S3A). In addition to the 2–340 region, CaM bound to the 2–21 fragment, but not to the fragments 30–340, 19–340 or 19–34 fragments (Fig. 3B, C), indicating that the first α-helix is responsible for the binding of CaM to the 2–340 INF2 fragment. Analysis of sequential deletions of the 2–21 sequence indicated that the 6–19 fragment includes the CaMBS (Fig. 3C).
CaM binds the N-terminal α-helix of INF2. A Schematic of the INF2 fragments used in the pull-down experiments. B, C Pull-down analysis of the association of GFP-CaM with GST fusions of the indicated INF2 fragments. D GST fusions of the fragment comprising amino acids 4–19 of INF2 with the indicated mutations in residues W11, L14 and L18 were used in pull-down experiments with GFP-CaM. The purified GST fusions were stained with Coomassie blue as a control for the amount of GST fusions used in (B–D). E Lysates of HEK293T cells expressing intact INF2 or INF2 with the triple W11L14L18A mutation fused to mCherry were immunoprecipitated with control or anti-Cherry antibodies and immunoblotted for exogenous INF2 with anti-Cherry antibodies, and with antibodies to endogenous CaM
We carried out a point-mutational analysis of the 6–19 peptide and found residue W11 to be essential for the binding of CaM, since its substitution by A or by any hydrophobic amino acid totally blocked CaM binding (Fig. 3D). Mutation of residues L14 and L18 to A diminished and suppressed, respectively, the binding of CaM. Whereas L18 tolerated substitution by any hydrophobic amino acid, although with reduced binding, the replacement of L14 by W or F, but not by V or I, abolished CaM binding (Fig. 3D). Individual substitution of each of the other residues by A did not significantly alter the binding of CaM (Fig. S3B, C). Mutation of A12 and A13 by Q did not affect the binding of CaM. In the case of A12, the substitution by E did not abolish, but did reduce, CaM binding despite the dramatic change in size and charge (Fig. S3C). The importance of the 2-19 INF2 sequence and the involvement of the three residues—W11, L14 and L18—was confirmed by pull-down assays with GST fused to CaM and cell extracts containing INF2 Δ2-18 or full-length INF2 proteins with single (W11A or L14A), double (L14L18A) or triple (W11L14L18A) mutations (Fig. S3D), and by co-immunoprecipitation analysis of intact INF2 and INF2 W11L14L18A with endogenous CaM (Fig. 3E). Equilibrium binding affinity measurements by surface plasmon resonance analysis of the interaction between a synthetic peptide covering the 2-19 sequence and purified bovine brain CaM yielded an apparent KD = 1.40 ± 0.48 μM (mean ± SD; 5 independent measurements; representative experiment in Fig. 4A). A similar value (KD = 1.2 ± 0.48 μM; 5 independent measurements; representative experiment in Fig. S3E) was obtained with CaM purified from bovine testes. A longer peptide encompassing amino acids 2–36 yielded a KD value in the same range as that of the 2-19 peptide, indicating that the INF2 sequence most proximal to the C-terminal of the 2-19 peptide has no effect on the affinity (Fig. S3F). The result of these experiments, which were done using purified CaM and synthetic peptides corresponding to the N-terminal extension of INF2, indicates that CaM interacts directly with INF2. The double L14L18A 2-19 peptide mutant displayed a reduced affinity with respect to the intact 2-19 peptide (Fig. 4B), whereas the interaction was undetectable for the single W11A and the triple W11L14L18A peptide mutants (Fig. 4C, D) and for the intact peptide in the absence of Ca2+ (Fig. 4E). In summary, the results in Figs. 3, 4 and S3 show that the site identified in the first helix of INF2 is the sole CaMBS detected in INF2, and that the affinity of the peptide–CaM interaction is within the low μM range.
Surface plasmon resonance analysis of the interaction of the 2-19 INF2 peptide with CaM. A–D Representative equilibrium binding affinity measurements of the interaction of purified CaM from bovine brain with the control 2-19 peptide (A, left panel) or 2-19 peptides containing the L14L18A (B), W11A (C) and W11L14L18A (D) mutations in the presence of Ca2+. E Control of the binding of CaM to the 2-19 peptide in the absence of Ca2+. The equilibrium plots of CaM with the control 2-19 (A, right panel) is shown, with indication of the apparent KD resulting from the same experiment. RU resonance units, Req RU at equilibrium
The CaMBS of INF2 binds the C-terminal lobe of CaM, and is conserved in vertebrates and in the human population
Phylogenetic analysis of the human 2-19 INF2 sequence in vertebrates shows the absolute conservation of residue W11 in all the species examined, whereas residues L14 and L18 are also maintained, except in the cases of conservative substitution of L14 by V in some reptiles, and of L14 and L18 by V in fishes (Figs. 5A and S4A). The analysis of the INF2 sequences deposited in public repositories indicated that, whereas most residues in the 6–19 peptide present multiple allelic forms in the human population, residues W11 and L18 are fully preserved and L14 shows only a low-frequency substitution by V (Figs. 5B and S4B). It is of note that all the pathogenic variants of INF2 except A13T are excluded from the N-terminal extension. This mutation, which was initially assigned as a mutation responsible for focal segmental glomerulosclerosis [10], was subsequently considered to be just a polymorphism with no pathological implications, since it is found at a significant frequency in the normal human population [9, 11], and behaves as normal INF2 when it is expressed in cultured cells, unlike the pathogenic INF2 forms, which are constitutively active [18]. In conclusion, these analyses indicate selective preservation of the most critical residues of the INF2 CaMBS.
Conservation of the CaMBS of INF2. A Alignment of the sequence of the 1–19 peptide of human INF2 with that of 50 species from different groups of vertebrates. The corresponding position of the W11, L14 and L18 residues of human INF2 is indicated. B Compilation of allelic variants of the 6–19 sequence of INF2 in the general human population, with indication of the SNP reference, database and frequency. C Examples of CaM-binding proteins with a sequence similar to the 1-4-8 motif of INF2. AKAP79 A-kinase-anchoring protein 79, CX32 connexin 32, GRM5 glutamate metabotropic receptor 5, PDE1A and PDE1B Ca2+/CaM-dependent 3´,5´-cyclic nucleotide phosphodiesterases 1A and 1B, RIT1 and RIT2 Ras-related GTPases 1 and 2, TRPV4 transient receptor potential vanilloid subtype 4
Canonical motifs for Ca2-dependent binding of CaM usually consist of an α-helix with two bulky, hydrophobic amino acids (W, F, I, L, V) at the first and last positions, spaced by a short stretch of a variable number of residues [25]. The motifs involved in Ca2+-dependent binding of CaM are classified according to the position of the hydrophobic amino acids involved in the binding [25, 26]. Our mutational and structural analyses indicated that the CaMBS of INF2 corresponds to a 1-4-8 motif in which W11 occupies the position 1 and L14 and L18 the positions 4 and 8, respectively. This type of motif has not been identified before, although it is present in other proteins known to bind CaM (Fig. 5C).
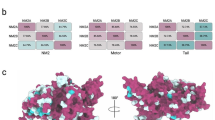
CaM contains four canonical EF hands that act as high-affinity Ca2+ binding motifs. Two of them are located at the N terminus (N-lobe), and the other two are situated at the C terminus (C-lobe), the two lobes being connected by an extremely flexible central linker region [27]. We used NMR to determine how CaM binds the 2-19 INF2 peptide. Upon titration with the 2-19 INF2 peptide the cross-peaks in the 1H,15N-HSQC spectra of CaM shifted (Fig. S4C), which confirmed that the 2-19 INF2 peptide binds CaM. The CaM residues experiencing the greatest changes were mainly located at the C-terminal lobe of CaM (Fig. S4D), indicating that the binding to the 2-19 INF2 peptide takes place through this domain. We considered the residues involved in the interaction, i.e., those identified in the 2-19 INF2 peptide, as being important for the binding (Fig. 3D), and those of CaM that underwent the largest chemical shift perturbation (Fig. S4D), and obtained a model of the complex (Fig. 6A) by docking the 2-19 INF2 structure (Fig. S1E) to a CaM structure (pdb code: 3CLN). The INF2 W11 residue appears trapped in a cleft formed by the two EF hands of the CaM C-lobe, which is the canonical binding position for the first hydrophobic anchor amino acid. That the C-terminal lobe of CaM mediates the binding was confirmed by pull-down experiments using GST fused to the INF2 2–21 peptide and GFP-CaM proteins with mutations that inactivated either the EF hands of the N-terminal or the C-terminal lobes, or all the EF hands simultaneously (Fig. 6B).
Structural model of the CaM/CaMBS complex. A Structural model of C-lobe of CaM bound to the 2-19 INF2 peptide. The position of the three hydrophobic amino acids comprising the 1-4-8 motif of the CaMBS of INF2 is indicated. B GST, alone or fused to the 2–21 peptide of INF2, was used in pull-down experiments with GFP fused to intact (WT) or to CaM with mutations that inactivate the two lobes (mutant 1234), the N-terminal lobe (mutant 12) or the C-terminal lobe (mutant 34). The purified GST fusions were stained with Coomassie blue as a control for the amount of GST protein used
The calmodulin-binding site of INF2 is crucial for INF2 activity
Once the CaMBS was identified, we analyzed how it modulates the functionality of INF2. Consistent with the relative importance of the W11, L14 and L18 residues in the 1-4-8 motif for CaM binding, the expression of intact INF2 efficiently restored the perinuclear F-actin ring and the cytosolic content of F-actin in INF2 KO cells, whereas the efficacy decreased gradually with INF2 L14A and INF2 L14L18A and was negligible with INF2 W11A and INF2 W11L14L18A (Fig. 7A–C). To analyze the effect of the inactivation of the CaMBS in the INF2 response to increased intracellular Ca2+ levels, we generated MDCK cells that stably expressed the fluorescent Ca2+ sensor GCaMP6S [28]. F-actin was stained in live cells with SiR-Actin to monitor the actin response of the cells to treatment with the calcium ionophore A23187. Since SiR-Actin is a fluorescent derivative of jasplakinolide that, at the concentrations used, might affect actin dynamics and stability [29], we only analyzed the initial burst of actin polymerization. Control and INF2 KO cells had similar levels of the sensor (Fig. S5A) and increased the fluorescent signal of the sensor upon treatment with A23187 (Fig. S5B). Consistent with previous results in other cell lines [16,17,18, 30], INF2 MDCK KO cells failed to increase actin polymerization upon A23187 treatment, whereas control cells responded rapidly (Fig. S5B, C and Video 1). The expression of INF2 W11L14L18A failed to rescue the defect in actin polymerization, whereas intact INF2 succeeded (Fig. 8A, B, and Video 1). As a control, we checked that INF2 W11L14L18A was not underexpressed relative to exogenous INF2-1 (Fig. S5D, E). The INF2 W11A and INF2 Δ18 mutants behaved similar to W11L14L18A, whereas the effect of INF2 L14A and INF2 L14L18A were similar to that of intact INF2, although they were less efficient (Fig. 8B). These results are consistent with the relative importance of the W11 and the two L residues of the 1-4-8 INF2 motif in the affinity of the 2-19 INF2 peptide for CaM (Fig. 4A–D).
Integrity of the CaMBS of INF2 is crucial for actin homeostasis. A–C INF2 KO cells expressing mCherry fusions of the indicated INF2 mutants were stained for F-actin. Nuclei were visualized with DAPI. An enlargement of the DAPI and F-actin staining corresponding to the boxed region is shown in the rightmost panels. Scale bar, 10 μm(A). Box plots showing the intensity of F-actin staining at the perinuclear region (B) and the cytosol (C) relative to that of control INF2 KO cells (> 150 cells were analyzed for each experimental condition; three independent experiments; ns not significant; *p > 0.05; ***p < 0.001)
The CaMBS of INF2 is required for actin polymerization and incorporation of GFP-MRTF-A into the nucleus in response to increased intracellular Ca2+ levels. A, B GCaMP6S INF2 KO cells expressing the indicated exogenous INF2 proteins were incubated with SiR-actin to visualize F-actin. Cells were treated with A23187 (0 min) and analyzed by videomicroscopy before and after treatment (A). Note that, although the staining of F-actin with SiR-Actin in live cells was poor compared to that of fluorescent phalloidin in fixed cells, SiR-Actin was useful for detecting the initial increase of the levels of F-actin after A23187 stimulation. B Kinetics of the perinuclear content of F-actin as measured by SiR-actin fluorescence relative to that of cells at t = 0 min (> 30 cells were analyzed for each experimental condition; three independent experiments). C–E GFP-MRTF-A INF2 KO cells expressing mCherry fusions of intact INF2 or the INF2 W11L14L18A mutant were treated with A23187 and analyzed by videomicroscopy before and after treatment (C). (D) Kinetics of the number of INF2 KO cells with nuclear distribution of GFP-MRTF-A relative to that of cells at t = 0 min (≥ 30 cells were analyzed for each experimental condition; three independent experiments). Scale bars, 5 μm. (E) The histogram shows the percentage of cells with GFP-MRTF-A predominantly in the nucleus after 5 min of treatment (> 190 cells were analyzed for each experimental condition; 3 independent experiments; ***p < 0.001). The mean ± SD is shown in ( B, D, E)
The myocardin-related transcription factor (MRTF) forms a complex with the serum response factor (SRF), a widely expressed transcription factor of mammals, and regulates its activity [31, 32]. At high G-actin concentrations in the cytosol, MRTF forms a reversible complex with G-actin and is held in an inactive state. At low G-actin concentrations, G-actin-free MRTF enters the nucleus and associates with SRF to direct gene transcription [33]. We previously reported that INF2 controls MRTF distribution, and consequently MRTF/SRF-mediated transcription, by modulating the cytosolic levels of free G-actin in human epithelial RPE-1 cells [34]. In HeLa cells, MRTF-A translocates to the nucleus in response to increased Ca2+ levels in an INF2 expression-dependent manner [17]. To compare the response of MRTF-A to increased intracellular Ca2+ levels in MDCK cells, we transiently expressed GFP-MRTF-A in control and INF2 KO cells and treated them with A23187 for 5 min. Unlike control cells that had GFP-MRTF-A predominantly in the nucleus, MRTF was mostly in the cytoplasm of INF2 KO cells (Fig. S5F, G). Then we used INF2 KO cells stably expressing GFP-MRTF-A to carry out rescue experiments and observed that intact INF2, but not INF2 W11L14L18A, allowed rapid incorporation of GFP-MRTF-A into the nucleus in response to A23187 treatment (Fig. 8C–E and Video 2). In conclusion, the results in Figs. 7 and 8 show that the integrity of the CaMBS of INF2 is necessary for INF2 function at steady state and in response to Ca2+ signaling.
Functional relevance of the calmodulin-binding site of INF2
In INF2, the direct DID–DAD interaction is weak [35], but is considerably strengthened by a complex of lysine-acetylated actin (KAc) and cyclase-associated protein (CAP) [36]. The in vitro interaction of CAP with the DID and of KAc with the DAD led to an inhibition model being proposed in which the complex serves as a bridge between the DID and the DAD [37]. The observation that increased intracellular Ca2+ levels produces a transient decrease in lysine acetylation of actin, led to the proposal that Ca2+ activates INF2 activation by decreasing the content of KAc and, thereby, reducing the levels of the CAP-Ac inhibitory complex [36]. mDia1 is activated by the binding of Rho to the GBD (G domain and DID) at the N-terminal region of the molecule (Fig. 9, top panel). Due to the slightly overlapping sites of Rho and DAD binding in mDia1, a two-step binding mechanism was postulated in which Rho makes preliminary contact with the G domain forming a loosely bound complex in a first step and, in a second step, interaction with the DID and steric interference and/or charge-charge repulsion lead to dissociation of the DAD [4, 7]. In our analysis of the 2–340 INF2 fragment, we only detected the CaMBS present in the first α-helix of the N-terminal extension. Therefore, it could be that the binding of CaM to this site is sufficient to promote a conformational change in the DID that releases the inhibitory interaction with the DAD mediated by the CAP-KAc complex (Fig. 9, bottom panel). However, our experiments cannot rule out the possibly existence of cryptic CaMBS sites in the region examined that could be unmasked by the binding of CaM to the CaMBS in the first α-helix. Although speculative at this stage, it is also plausible that, by analogy with the two-step mechanism proposed mDia1 activation [4, 7], the binding of the C-terminal lobe of CaM to the CaMBS present in the N-terminal extension of INF2 exposes a cryptic CaMBS in the DID that binds CaM, perhaps through the free N-terminal lobe of the CaM bound to the first α-helix. The binding of CaM to this site would disrupt the DID–CAP interaction, breaking the inhibitory bridge between the DID and the DAD formed by the CAP-KAc complex. Whichever mechanism operates, it would work in concert with decreased KAc levels to rapidly activate INF2 in response to the increased intracellular Ca2+concentration [36].
Ca2+ is a central second messenger of signal transduction pathways in eukaryotic cells. CaM, which is ubiquitously expressed, acts as a signaling molecule translating transient fluctuations of Ca2+ levels inside cells into rapid and appropriate responses [38, 39]. CaM normally constitutes > 0.1% of the total protein present in cells (0.1–1.0 μM) [40], and so does not limit the interaction with INF2. The concentration of intracellular free Ca2+ (approximately 0.1 μM) of most cells and the affinity of CaM for Ca2+ (KD = 0.5–5.0 μM) [40, 41] imply that only a small fraction of CaM is in the form of Ca2+/CaM at steady state. According to our results, this pool would be the responsible for the basal activation of INF2 required to maintain the perinuclear F-actin ring and the cytosolic F-actin content that we observe under our cell culture conditions (Figs. 2 and S2A–C). Considering the affinity of the interaction of CaM with the CaMBS-containing INF2 peptide, only a substantial increase in intracellular Ca2+ levels—for example that induced by the application of mechanical force or treatment with physiological ligands (e.g., thrombin, lysophosphatidic acid) for G protein-coupled receptors or with calcium ionophore—would be able to mediate Ca2+/CaM binding to INF2 for rapid remodeling of the F-actin. This is consistent with previous observations [17, 18, 24, 36] and with our results (Fig. 8A, B).
In summary, we have determined the structure of the N-terminal extension of INF2 and found it to be organized into two α-helices, the first of which contains a Ca2+-dependent CaMBS in which the hydrophobic residues involved in the binding are arranged into a 1-4-8 motif that is conserved in vertebrates and in the human population. The identification of the INF2 CaMBS will allow the use of mutants with this site inactivated to distinguish the events mediated by Ca2/CaM involving INF2 activation from those that are INF2 independent. Considering the relevance and ubiquity of Ca2/CaM signaling, the remodeling of the cytoskeleton by Ca2/CaM-mediated INF2 activation, its reported influence on gene expression, and the conservation of the INF2 CaMBS, it is likely that the INF2 CaMBS modulates multiple signaling pathways and physiological processes in vertebrates [17, 24, 42,43,44,45].
Materials and methods
Materials
SOURCE | IDENTIFIER | |
|---|---|---|
Antibodies | ||
Mouse monoclonal anti-CaM | Millipore | Cat#05–173 |
Rabbit polyclonal anti-Cherry | Abcam | Cat#ab167453 |
Mouse monoclonal anti-GADPH (clone 6C5) | Ambion | Cat#AM4300 |
Mouse monoclonal anti-GFP | Sigma-Aldrich | Cat#11814460001 |
Rabbit IgG control antibody, unconjugated | Sigma-Aldrich | Cat#I5006 |
Rabbit polyclonal anti-INF2 | N/A | |
Secondary donkey anti-mouse, Alexa Fluor 488 conjugated | Thermo Fisher | Cat#A-21202 |
Secondary donkey anti-rabbit IgG, HRP conjugated | GE Healthcare | Cat#NA934 |
Secondary donkey anti-mouse IgG, HRP conjugated | Jackson ImmunoResearch Labs | Cat#715–035-151 |
Bacterial strains | ||
E. coli BL21 | Stratagene | Cat#200131 |
E. coli XL10-Golda | Stratagene | Cat#200314 |
E. coli Rosetta 2(DE3)b | Merck | Cat#71397 |
Chemicals, peptides, and proteins | ||
A23187 | Sigma-Aldrich | Cat#C7522 |
Alexa Fluor™ 488 Phalloidin | Thermo Fisher | Cat#A12379 |
Calmodulin from bovine brain | Sigma-Aldrich | Cat#208694 |
Calmodulin from bovine testes | Sigma-Aldrich | Cat#P1431 |
DAPI | Merck | Cat#268298 |
Fetal bovine serum | Gibco | Cat#A4766801 |
Formalin solution, neutral buffered, 10% | Sigma-Aldrich | Cat#HT501128 |
Fluoromount | Merck | Cat#F4680 |
G418 sulfate | ChemCruz | Cat#108321–42-2 |
Glutathione Sepharose® 4B | Cytiva | Cat#GE17-0756–01 |
Polybrene | Sigma-Aldrich | Cat#TR-1003-G |
Polyethylenimine | Polysciences | Cat#23966 |
Protease inhibitor cocktail | Merck | Cat#11697498001 |
P rotein A-Sepharose® CL-4B | Cytiva | Cat#GE17-0780–01 |
Sample buffer, Laemmli 2 × concentrate | Sigma-Aldrich | Cat#S3401 |
SiR-Actin | Spirochrome | Cat#SC001 |
Surfactant P20 | Cytiva | Cat#BR-1000–54 |
Triton X-100 | Merck | Cat#9036–19-5 |
T ween-20 | Sigma-Aldrich | Cat#9005–64-5 |
Peptide INF2 2-19c (KGGSVKEGAQRKWAALKEKLG) | Proteomics Facility (CNB, CSIC) | N/A |
Peptide INF2 2-19 W11Ac KGGSVKEGAQRKAAALKEKLG | Proteomics Facility (CNB, CSIC) | N/A |
Peptide INF2 2-19 L14L18Ac KGGSVKEGAQRKWAAAKEKAG | Proteomics Facility (CNB, CSIC) | N/A |
Peptide INF2 2-19 W11L14L18Ac KGGSVKEGAQRKAAAAKEKAG | Proteomics Facility (CNB, CSIC) | N/A |
Peptide INF2 2-36c KGGSVKEGAQRKWAALKEKLGPQDSDPTEANLESADPE | pepMic Co., Ltd | N/A |
Peptide INF2 2-36d SVKEGAQRKWAALKEKLGPQDSDPTEANLESADPE | CASLO ApS | N/A |
Peptide INF2 2-19d SVKEGAQRKWAALKEKLG | CASLO ApS | N/A |
Commercial assays | ||
Lipofectamine 2000 | Thermo Scientific | Cat#11668019 |
QuikChange II Site-Directed Mutagenesis Kit | Stratagene | Cat#200523 |
Wizard® Plus SV Minipreps DNA Purification Systems | Promega | Cat#A1330 |
Deposited data | ||
Chemical shifts of INF2 2-19 in 30% trifluoroethanol | This paper | BMRB-51388 |
Chemical shifts of INF2 2-19 in aqueous solution | This paper | BMRB-51407 |
Chemical shifts of INF2 2-19 in 30% trifluoroethanol | This paper | BMRB-51408 |
Chemical shifts of CaM in aqueous solution | This paper | BMRB-51422 |
Chemical shifts of INF2 2-19 in 30% trifluoroethanol | This paper | BMRB-51388 |
Cell lines | ||
MDCK II | ATCC | Cat#CRL-2936 |
HEK293T | ATCC | Cat#CRL-3216 |
Recombinant DNA | ||
pEGFP-hCaM | Addgene | Cat#47602 |
pCaM12(D20A/D56A)/pIRES2-eGFP | Addgene | Cat#111512 |
pCaM34(D93A/D129A)/pIRES2-eGFP | Addgene | Cat#111517 |
pCaM1234(D20A/D56AD93A/D129A)/pIRES2-eGFP | Addgene | Cat#111518 |
pEGFP-C1 | Clontech | Cat#6084–1 |
pEGFP-CaM12 | This paper | N/A |
pEGFP-CaM34 | This paper | N/A |
pEGFP-CaM1234 | This paper | N/A |
pGP-CMV-GCaMP6S | Addgene | Cat#40753 |
pCMS28Cherry | Agromayor et al. [47] | N/A |
p33Cherry | This paper | N/A |
p3xFLAG-MKL1 | Addgene | Cat#11978 |
pEGFP-MRTF-A | This paper | N/A |
pEGFP INF2-1 | Andrés-Delgado et al. [46] | N/A |
pEGFP INF2-2 | Andrés-Delgado et al. [46] | N/A |
p33Cherry INF2-1 | This paper | N/A |
p33Cherry INF2-2 | This paper | N/A |
p33Cherry INF2-1 Δ2-18 | This paper | N/A |
p33Cherry INF2-1 Δ2-30 | This paper | N/A |
p33Cherry INF2-1 W11A | This paper | N/A |
p33Cherry INF2-1 L14A | This paper | N/A |
p33Cherry INF2-1 L14L18A | This paper | N/A |
p33Cherry INF2-1 W11L14L18A | This paper | N/A |
MLV-GagPol/pHIV 8.1 | Agromayor et al. [47] | N/A |
pHIT VSVg | Agromayor et al. [47] | N/A |
pGEX-4 T-1 | Cytiva | Cat#GE28-9545–49 |
pGEX-4T-1 INF2 2-340 | Andrés-Delgado et al. [46] | N/A |
pGEX-4EX | This paper | N/A |
pGEX-4EX INF2 30-340 | This paper | N/A |
pGEX-4EX INF2 19-340 | This paper | N/A |
pGEX-4EX INF2 19-34 | This paper | N/A |
pGEX-4EX INF2 2-21 | This paper | N/A |
pGEX-4EX INF2 4-19 | This paper | N/A |
pGEX-4EX INF2 6-19 | This paper | N/A |
pGEX-4EX INF2 8-19 | This paper | N/A |
pGEX-4EX INF2 4-19 G6R | This paper | N/A |
pGEX-4EX INF2 4-19 A7R | This paper | N/A |
pGEX-4EX INF2 4-19 Q8A | This paper | N/A |
pGEX-4EX INF2 4-19 R9A | This paper | N/A |
pGEX-4EX INF2 4-19 K10A | This paper | N/A |
pGEX-4EX INF2 4-19 W11A | This paper | N/A |
pGEX-4EX INF2 4-19 W11F | This paper | N/A |
pGEX-4EX INF2 4-19 W11L | This paper | N/A |
pGEX-4EX INF2 4-19 W11V | This paper | N/A |
pGEX-4EX INF2 4-19 W11I | This paper | N/A |
pGEX-4EX INF2 4-19 A12E | This paper | N/A |
pGEX-4EX INF2 4-19 A12Q | This paper | N/A |
pGEX-4EX INF2 4-19 A13E | This paper | N/A |
pGEX-4EX INF2 4-19 A13Q | This paper | N/A |
pGEX-4EX INF2 4-19 A13T | This paper | N/A |
pGEX-4EX INF2 4-19 L14A | This paper | N/A |
pGEX-4EX INF2 4-19 L14W | This paper | N/A |
pGEX-4EX INF2 4-19 L14F | This paper | N/A |
pGEX-4EX INF2 4-19 L14V | This paper | N/A |
pGEX-4EX INF2 4-19 L14I | This paper | N/A |
pGEX-4EX INF2 4-19 K15A | This paper | N/A |
pGEX-4EX INF2 4-19 E16A | This paper | N/A |
pGEX-4EX INF2 4-19 K17A | This paper | N/A |
pGEX-4EX INF2 4-19 L18A | This paper | N/A |
pGEX-4EX INF2 4-19 L18W | This paper | N/A |
pGEX-4EX INF2 4-19 L18F | This paper | N/A |
pGEX-4EX INF2 4-19 L18V | This paper | N/A |
pGEX-4EX INF2 4-19 L18I | This paper | N/A |
pGEX-4EX INF2 4-19 G19A | This paper | N/A |
R701-X65-527: His6-tev-Hs.CALM1(2-148) | Addgene | Cat#159693 |
Software | ||
Adobe Illustrator | Adobe | |
BIAevaluation Software | Biacore AB | |
Fiji | Open Source | |
gnomAD | Broad Institute | |
Microsoft Excel | Microsoft | |
NCBI SNP | NCBI | |
NHLBI EVS | NIH NHLBI | |
R Studio | Open Source | |
TOPMed | NIH NHLBI | |
Topspin | Bruker | https://www.bruker.com/en/products-and-solutions/mr/nmr-software/topspin.html |
NMRPipe | Delaglio et al. [48] | |
MddNMR | Orekhov et al. [49] | |
NMRFAM-Sparky | UCSF/ Univ. Madison-Wisconsin | |
Talos-N | NIH NIDDK | |
Cyana 3.98 | P. Guntert | http://www.bpc.uni-frankfurt.de/guentert/wiki/index.php/Software |
Molmol | Koradi et al. [50] | |
Haddock 2.4 | Univ. Utrecht | |
Other | ||
Sensor Chip SA | Cytiva | BR100032 |
µ-Slide 8 Well | Ibidi | Cat#80826 |
Cells and cell culture
Epithelial canine MDCK II cells, MDCK II INF2 KO cells [20] and human epithelial HEK293T cells were grown in MEM supplemented with 5% or 10% (v/v), respectively, fetal bovine serum (FBS) at 37 °C in a 95% air/5% CO2 atmosphere. Mycoplasma testing was regularly performed.
Molecular cloning
The polylinker of the GST vector pGEX-4T1 was modified by insertion of the polylinker sequence 5´-GAATTCTCCGGAGGCGGCCGCTCTAGACTCGAG-3´, using annealed oligonucleotides, generating the pGEX-4EX plasmid. The inserted sequence contains one BspEI site and one XhoI site at the 5´ and 3´ end, respectively, that were used for cloning. For expression in E. coli as GST fusions, the INF2 DNA fragments 18–340 and 30–340 were obtained by PCR using the appropriate forward and reverse oligonucleotide primers with a BspEI site (forward primer) and a XhoI site (reverse primer) at their 5´ end, and INF2-1 cDNA as the template. The products were cloned between the BspEI and XhoI sites of pGEX-4EX. For the human INF2 2–21, 4–19, 6–19 and 8–19 GST constructs and derivative mutants, the appropriate forward and reverse oligonucleotide with preformed 5´ BspEI (forward oligonucleotide) and XhoI (reverse oligonucleotide) cohesive ends were annealed and cloned between the BspEI and XhoI sites of pGEX-4EX. For the pull-down experiments with GST-CaM, we generated a DNA construct by cloning the XhoI/BamHI fragment, which contains the CaM coding sequence, from pEGFP-hCaM (Addgene #47602) in pGEX-4T1.
For mammalian expression, the polylinker of the retroviral pCMS28Cherry vector [47] was modified by deletion of the XhoI–BamHI fragment, which contains an internal ribosomal entry site and the puromycin resistance gene, elimination of the unique BamHI and SalI sites, and insertion of the polylinker sequence used to generate pGEX-4EX, to give rise to the p33Cherry vector. For expression as mCherry fusions, the entire coding sequence of human INF2-1 and INF2-2 obtained from pEGFP-C1 INF2-1 and pEGFP-C1 INF2-2 was excised with BspEI and XhoI and cloned in-frame of the mCherry coding sequence from p33Cherry. The p33Cherry INF2-1 Δ2–18 and Δ2–30 constructs were obtained using the BspEI–SalI DNA fragment from pGEX-4EX INF2 18–340 and 30–240 constructs, respectively, to replace the corresponding BspEI–SalI fragment of p33Cherry INF2-1, using the unique SalI site present in the sequence encoding the DID of human INF2. For the W11A, L14A, L14L18A and W11L14L18A INF2 mutants in p33Cherry the INF2 fragment used was generated by digestion with BspEI and SalI of PCR products, obtained using forward and reverse primers with the appropriate modifications, and INF2-1 as template.
The constructs pEGFP-CaM12, 34 and 1234 were obtained by replacing the XhoI–BamHI CaM insert in pEGFP-hCaM with the corresponding XhoI–BamHI CaM DNA fragment obtained from pCaM12(D20A/D56A)/pIRES2-eGFP, pCaM34(D93A/D129A)/pIRES2-eGFP and pCaM1234(D20A/D56AD93A/D129A)/pIRES2-eGFP [51]. pGFP-MRTF-A was obtained by in-frame cloning the MRTF-A EcoRI–BamHI insert from p3xFLAG-MKL1 into pEGFP-C1. All constructs were verified by DNA sequencing (Macrogen).
Retroviral infections
To pack retroviral plasmids into retroviral particles, HEK293T cells in a p100 culture dish were cotransfected with p33Cherry constructs expressing the indicated mCherry-INF2 fusions and the plasmids MLV-GagPol/pHIV 8.1 and pHIT VSVg (1.4 μg/ml) in a 4.6:3.3:1 ratio using polyethyleneimine [47]. After 48 h the supernatant containing the viral particles was collected and filtered. For expression of mCherry alone or mCherry-fused INF2 proteins, MDCK cells were treated with 10 μg/ml polybrene at 37ºC for 15 min and incubated with the supernatant containing the viral particles for 7 h. After that, cells were washed and incubated for 48 h.
Transfections and generation of stable transfectants
MDCK or MDCK KO cells stably expressing exogenous proteins (GCaMP6S or GFP-MRTF-A) were generated by transfection using Lipofectamine 2000 according to the manufacturer’s instructions. 48 h later, single cells expressing GFP were FACS sorted. In the case of MDCK INF2 KO cells stably expressing GFP-MRTF-A, but not in those expressing GCaMP6S, transfected cells were selected with 1 mg/ml G-418 for 14 days. Individual clones were screened by fluorescence microscopy and immunoblotting.
Confocal microscopy and videomicroscopy
Cells were fixed with 10% formalin (37% formaldehyde solution) and permeabilized with 0.2% Triton X-100 for 5 min on ice. After blocking with 3% (wt/vol) BSA for 30 min, cells were stained with phalloidin-Alexa 488 and DAPI, and then washed extensively. Coverslips were mounted on glass slides with Fluoromount. Fluorescence was examined with a Nikon A1R + confocal laser-scanning microscope with 60x (NA 1.2) water objective. Brightness and contrast were optimized with Fiji software (National Institutes of Health, USA). LSM images were converted to TIFF format. The levels of perinuclear F-actin were measured using a ROI consisting of a 0.5-μm-width ring around the nucleus, while those of cytosolic F-actin were measured in a 20 μm2 square ROI close to the central region of the cell. The data represented correspond to the average intensity of three planes. The images obtained were processed using Fiji (ImageJ). We used videomicroscopy to monitor actin dynamics using SiR-Actin, which is a fluorogenic reagent that allows labeling of F-actin in live cells [29]. SiR-actin emits in far-red because of its silicon rhodamine moiety, thereby making it compatible with the emission of the GFP (GCaMP6S) and Cherry (INF2) proteins expressed in our experiments. SiR-Actin (0.6 μM) was added to cells expressing GCaMP6S 1 h before recording. Verapamil (10 μM), an efflux pump inhibitor, was added simultaneously to increase the otherwise very low staining efficiency of MDCK cells. These cells or cells expressing GCaMP6S or GFP-MRTF-A were plated onto µ-Slide 8 well dishes, and maintained at 37 °C in MEM without phenol red supplemented with 12.5 mM HEPES, pH 7.0, and 5% (v/v) FBS during the recording. To increase the intracellular Ca2+ levels, cells were treated with 8 μM A23187 (time = 0). Cells were filmed with an Olympus Spinning Disk SpinSR10 with 60x (NA 1.3) silicone immersion objective. Brightness and contrast were optimized with ImageJ (https://imagej.nih.gov/ij/). Quantifications were performed using ImageJ software.
GST pull-down assay
For use in pull-down assays, GST-fused proteins were expressed in E. coli BL21 cells. Cells were grown in LB media at 37 °C until the culture reached an OD600 of 0.6–0.8, whereupon the temperature was decreased to 20 °C and 0.5 mM IPTG was added to induce expression of the recombinant protein for 16 h. Cell lysates of HEK293T cells expressing GFP-CaM or mCherry-INF2 proteins were incubated in assay buffer (PBS supplemented with 1 mM DTT, 0.1 mM PMSF, 0.2% Tween-20, 0.5 mg/ml DNase I, 5 mM Ca2+, and a commercial cocktail of protease inhibitors) at 4 ºC with 30 µg of the indicated GST-fused proteins immobilized on glutathione-Sepharose beads. After 3 h, the beads were collected by centrifugation in a microfuge at 800 rpm for 5 min, and washed twice with cold PBS. Samples were analyzed by immunoblotting. An aliquot from each glutathione-Sepharose bead-immobilized GST-fused protein was stained with Coomassie blue to control for the amount of GST protein used.
Immunoblotting
After blocking with 5% BSA (w/v) and 0.05% (v/v) Tween-20 in Tris-buffered saline, membranes were incubated overnight with the indicated primary antibodies, washed with Tris-buffered saline containing 0.05% Tween-20, and incubated for 30 min with the corresponding secondary antibodies coupled to HRP. The primary antibody used to detect INF2 recognizes both endogenous canine INF2 and exogenous human mCherry-INF2 [15, 46]. The signal was visualized with Clarity Western ECL substrate (BioRad).
Immunoprecipitation analysis
HEK293T cells expressing mCherry fusions of INF2 or INF2 W11L14L18A mutant were lysed at 4 ºC in 1 ml lysis buffer containing 20 mM Tris, pH 8.0, 150 mM NaCl, 5 mM CaCl2, 1.15% (v/v) glycerol, 1% (v/v) Nonidet P-40, 1 mM sodium orthovanadate, 0.1 mM phenylmethylsulphonyl fluoride, and a commercial cocktail of protease inhibitors. The cell extracts centrifuged in a microfuge at 14,000 rpm for 5 min, and the cleared supernatant was then incubated with control or anti-Cherry rabbit antibodies bound to protein A-Sepharose beads for 3 h. After four washing steps, bound proteins were eluted in Laemmli’s buffer and processed for immunoblotting to detect exogenous INF2 with anti-Cherry antibodies and endogenous CaM with anti-CaM antibodies.
Synthetic peptides
For use in surface plasmon resonance analysis, the peptide corresponding to amino acids 2-19 of human INF2 and similar peptides with replacements in the W11, L14 and L18 residues were synthesized in an automated multiple peptide synthesizer (Multipep, Intavis, Köln, Germany) in the Proteomics Facility of the Centro Nacional de Biotecnología (CSIC, Madrid). The INF2 2–36 peptide was obtained as a bespoke product with the same modifications from pepMic (Suzhou, China). These peptides contained two G residues at its N terminus to separate the INF2 sequence from an additional N-terminal end K residue, which was biotinylated in the ε-amino group. For use in NMR studies, the INF2 2–36 and 2-19 peptides were synthesized to order, without any modification, by CASLO ApS (Denmark). All the peptides were synthesized using the solid-phase synthetic procedures and standard Fmoc [N-(9-fluorenyl)methoxycarbonyl] chemistry [52], and then underwent reverse-phase HPLC purification.
Surface plasmon resonance analysis
All surface plasmon resonance experiments were carried out at 25ºC in a Biacore 3000 (Cytiva) robotic biosensor. Analyses were performed using HBSP (10 mM HEPES pH 7.4, 150 mM NaCl, 0.005% surfactant P20) supplemented with 1 mM CaCl2 as running buffer and for the dilution of analytes and ligands. Biotinylated peptides were individually immobilized on a streptavidin-coated sensor chip in flow cells 2, 3 and 4 at a flow rate of 10 µl/min (20–50 RU captured). The empty flow cell 1 was used as a control for non-specific binding and bulk effects. Purified CaM was injected at different concentrations ranging from 13.2 nM to 9.6 µM in duplicate, at a flowrate of 50 µl/min, with an association time of 60 s followed by dissociation for 60 s. After that, the sensor chip was regenerated by injection of 10 mM glycine–HCl, pH 1.7, with a contact time of 12 s. The signal obtained in the reference cell was subtracted from each of the sample sensorgrams and, afterwards, the signal corresponding to the injection of the buffer was also subtracted. Sensorgrams with different concentrations of CaM were overlaid, aligned and analyzed with BIAevaluation Software 4.1, using the double-reference method [53]. In all sensorgrams, the binding reached the equilibrium very quickly and the complete dissociation was achieved in a few seconds, due to the very fast association (Ka) and dissociation (Kd) constant rates, which were beyond the detection limits of the equipment. Therefore, we used the response signal at equilibrium at each concentration of CaM to calculate the affinity constant (KD). A Langmuir binding isotherm was fitted to determine KD using the equation: Req = C × Rmax/C + KD, where C is the analyte concentration and Rmax is the maximal binding capacity.
Expression and purification of CaM
For NMR spectroscopy, recombinant human CaM was produced using the plasmid R701-X65-527: His6-tev-Hs.CALM1(2–148) [54] expressed in E. coli Rosetta 2(DE3) cells. Isotopically labeled CaM was produced in supplemented minimal growth medium containing 15NH4Cl and/or 13C-glucose as the sole sources of nitrogen and carbon respectively, as previously described [55]. Briefly cells were grown in 2 L of LB media at 37 °C until and OD600 of 0.8 was obtained, whereupon they were centrifuged at 3000×g for 20 min and the pellet was resuspended in 0.5 ml of prewarmed, isotopically labeled minimal medium. Cells were incubated at 37 °C for 1–2 h, and the temperature was then decreased to 20 °C before induction with 0.5 mM IPTG. The cells were harvested after 16 h. For CaM purification, the pellet obtained by centrifugation was lysed by mechanical disruption through sonication and the supernatant was loaded into a nickel-based HisTrap FF Crude column (GE Healthcare) using buffer A (50 mM Tris–HCl pH 8.0, 300 mM NaCl and 10 mM imidazole). Protein was eluted with an imidazole gradient in buffer A from 10 mM up to 250 mM. Fractions containing CaM were dialyzed against buffer B (20 mM Tris–HCl pH 8.0, 10 mM NaCl) and digested with TEV-protease produced in-house using the protocol of Blommel and Fox [56]. The extent of the digestion was confirmed by the difference in the size of noncleaved and cleaved CaM, as assessed by SDS-PAGE. After removing the remaining uncleaved protein in a HisTrap FF Crude column, the cleaved product was buffer exchanged using a Hi-Prep desalting column (GE Healthcare) into the final buffer (20 mM Tris–HCl pH 8.0, 10 mM NaCl and 5 mM CaCl2 pH 7.0). Eluted fractions containing CaM were concentrated to an appropriate volume and the sample concentration was estimated using a Nanodrop spectrophotometer. The identity of CaM was confirmed by MALDI-TOF mass spectrometry.
NMR spectroscopy of peptides
The deuterated compounds [D3]-2,2,2-trifluoroethanol (TFE) (99%) and D2O (99.9%) were purchased from Eurisotop, France. Peptide samples for NMR spectra acquisition were prepared at 1.0 mM concentration in a final volume of 500 μl of either H2O/D2O (9:1 v/v) or 30% [D3]-TFE/70% H2O/D2O (9:1 v/v). The pH was measured with a glass microelectrode and adjusted to the desired value of 5.5 by adding NaOD or DCl and was not corrected for isotope effects. All samples contained 1.5 µl of sodium 2,2-dimethyl-2-silapentane-5-sulfonate (DSS) as an internal reference for 1H chemical shifts. Samples were placed in 5-mm NMR tubes. 1H 1D and 2D spectra of peptides were recorded on a Bruker AVNEO-600 spectrometer operating at a proton frequency of 600.13 MHz and equipped with a cryoprobe. The acquired 2D spectra were: 1H,1H double-quantum filtered correlation spectroscopy (COSY), 1H,1H total correlation spectroscopy (TOCSY), 1H,1H nuclear Overhauser effect spectroscopy (NOESY) and 1H–13C heteronuclear single quantum coherence (HSQC) spectra at 13C natural abundance. The water signal was suppressed using an excitation sculpting scheme [57]. All spectra were processed using Topspin 4.0.8 program. Baseline correction was applied in both dimensions. 13C δ-values were indirectly referenced using the IUPAC-IUB recommended 1H/13C chemical shift ratio (0.25144953) [58]. 1H and 13C chemical shifts for the peptides were assigned by analyzing the 2D NMR spectra and with the aid of the NMRFAM-SPARKY software [59]. The chemical shift values were deposited in BioMagResBank under accession codes 51,388–51,389 for peptide INF2 2-19 and 51,407–51,408 for peptide INF2 2–36.
NMR spectroscopy of CaM
Protein samples for NMR spectrum acquisition were prepared by adding 50 μl D2O to a 450 μl of protein in the final buffer (20 mM Tris–HCl, pH 7.0; 10 mM NaCl; 5 mM CaCl2). Protein concentration was approximately 250 μM. 2D 1H,15 N-HSQC and series of 3D spectra NHCO, HN(CA)CO, HNCA, HN(CO)CA, HNCACB, and CBCA(CO)NH were acquired using 13C,15 N-CaM samples on a Bruker AVNEO-800 spectrometer operating at proton frequencies 800.1 MHz, and equipped with a TCI cryoprobe. The acquisition parameters for these spectra are presented in Table S2. All spectra were processed using Topspin 4.0.8 program A methanol sample was employed to calibrate cryoprobe temperature. 1H, 15 N and 13C chemical shifts for backbone atoms and for Cβ carbons of free CaM and for the CaM/INF2-2-19 complex were assigned by analysis of the series of acquired 3D spectra with the aid of the NMRFAM-SPARKY software [59]. The chemical shift values were deposited in BioMagResBank under accession code 51,422.
Determination of secondary structure
The elements of the secondary structure of peptides and proteins can be determined from the ΔδHα and ΔδCα values, which are the differences in the chemical shifts of 1Hα protons and 13Cα carbons from reference values characteristic of random coil. Thus, ΔδHα = δHαobserved – δHαRC, ppm, and ΔδCα = δCαobserved – δCαRC, ppm, where δHαobserved and δCαobserved are the observed chemical shifts for 1Hα and 13Cα nuclei, respectively, and δHαRC and δCαRC are reference random coil values for the 1Hα and 13Cα chemical shifts, respectively. These reference values were taken from Wishart et al. [60]. α-helices are identified by stretches of negative ΔδHα and positive ΔδCα values, and extended/β-strand segments by positive ΔδHα and negative ΔδCα values. Values within the ranges − 0.04 ppm < ΔδHα < + 0.04 ppm and − 0.4 ppm < ΔδCα < + 0.4 ppm are considered to indicate random coil. The helix populations can be estimated from the ΔδHα values averaged for the helical residues—amino acids 3–17 in the case of the N-terminal helix of the INF2 peptides—divided by the ΔδHα value corresponding to 100% helix (− 0.39 ppm), that is, % helix = 100 × ⟨ΔδHα⟩/(− 0.39) [61]. Thus, the populations of the N-terminal helix formed by both peptides are approximately 23% in aqueous solution, increasing to 43–47% in the presence of TFE. Assuming an experimental error of ± 0.01 ppm in the measurement of 1H chemical shifts, the error in the estimated populations would be ± 3%.
Peptide structure calculation
Peptide structures were calculated using the standard iterative protocol for automatic NOE assignment of the CYANA 3.98 program [62], which performs seven cycles of combined automated NOE assignment and structure calculation, being 100 conformers calculated per cycle. The experimental input data were the lists of: (1) assigned chemical shifts, (2) NOE integrated cross-peaks present in 150 ms NOESY spectra, and (3) ϕ and ψ dihedral angle restraints. The automatic integration subroutine of SPARKY software (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco) was used to integrate NOE cross-peaks. Restraints for dihedral angles were obtained from 1H and 13C chemical shifts using the TALOSn webserver [63]. The final structure of each peptide is the ensemble of the 20 lowest target function conformers calculated in the final cycle. Table S1 provides a summary of the statistical parameters of these structural ensembles, which were visualized and examined using the MOLMOL program [50]. Structures are available upon request from the authors.
Titration of CaM with INF2-2-19 peptide
To determine experimentally how CaM and the N-terminal region of INF2 interact, a 15 N-CaM sample was titrated with increasing quantities of INF2-2-19 peptide. 2D [1H-15 N]-HSQC spectra were acquired at 25 °C with protein/peptide ratios of 1:0, 1:0.1, 1:0.2, 1:0.5, 1:1, 1:2, 1:4 and 1:8. Since most cross-peaks in the [1H-15 N]-HSQC spectra were shifted upon peptide titration (Fig. S3C), to confirm cross-peak assignment a 13C,15N-CaM/INF2-2-19 sample at 1:1 ratio was used to acquire a series of 3D spectra (HNCA, HN(CA)CO, HN(CO)CA, CBCANH, CBCA(CO)NH and NHCO), which were acquired using non-uniform sampling (NUS) and were processed using NMRPipe [48] and MddNMR software (http://mddnmr.spektrino.com) [49]. Details of the acquisition parameters of these spectra are presented in Table S2. Analysis of these spectra enabled the almost complete assignment of 1H, 15 N, and 13C backbone resonances of CaM in its complex with the INF2 2-19 peptide. Chemical shift perturbations (CSPs) upon peptide titration were calculated using the equation CSP = [(Δδ1H)2 + (Δδ15N/5)2 × 0.5]1/2, in which Δδ1H and Δδ15N are the chemical shift changes in the amide 1H and 15 N resonances, respectively. Using the Schumann criteria [64], the threshold for significant changes was 0.25 ppm.
NMR-driven docking (HADDOCK)
To visualize how CaM–INF2 interaction occurs, a complex model was built using the Haddock webserver [65, 66]. This program requires input coordinates for both, protein and peptide, and a definition of their interaction residues, classified as active or passive. Protein coordinates were taken from pdb code 3CLN. Peptide coordinates were those of the lowest target function conformer of the structural ensemble calculated in this work. The active residues for the INF2-2-19 peptide were those determined by the pull-down analysis carried out in this work, and those for the protein were the ones exhibiting the greatest chemical shift perturbations (CSP; Δδav > 0.25 ppm; see Fig. S3D). Representative complexes were those with the best Haddock docking scores. Figure 6A shows the structure with the lowest Haddock docking score.
Bioinformatic analyses
The protein encoded by the INF2 gene in the Human Genome Resources at the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/genome/guide/human) was taken as the human INF2 reference for sequence comparisons. NCBI blast was used to align the human INF2 sequence with that of the indicated vertebrates (Fig. 5A). INF2 sequences from more than 300 different vertebrates were analyzed in Fig. S3A. Only the variants found in at least two different species were considered. The following databases were searched to find INF2 variants in the human population: gnomAD, LOVD3, NCBI Clin Var, NCBI SNP, NHLBI EVS and TOPMed (Fig. 5B). The CaM-binding protein database (Gene Ontology, GO:0,005,516) and the CaM target database (http://calcium.uhnres.utoronto.ca/ctdb/ctdb) were interrogated to identify proteins with a Ca2/CaM binding motif similar to the INF2 1-4-8 motif.
Statistical analysis
Most graphs and statistical analysis were performed with RStudio, version 1.2.5033 software. Statistical significance of differences between means was assessed with two-tailed Student’s unpaired t-tests, while differences between median values were examined using the Mann–Whitney–Wilcoxon test, as indicated. Additional information is provided in the figure legends.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon request.
References
Chesarone MA, DuPage AG, Goode BL (2010) Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Biol 11:62–74. https://doi.org/10.1038/nrm2816
Goode BL, Eck MJ (2007) Mechanism and function of formins in the control of actin assembly. Annu Rev Biochem 76:593–627. https://doi.org/10.1146/annurev.biochem.75.103004.142647
Campellone KG, Welch MD (2010) A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Biol 11:237–251. https://doi.org/10.1038/nrm2867
Lammers M, Meyer S, Kühlmann D, Wittinghofer A (2008) Specificity of interactions between mDia isoforms and Rho proteins. J Biol Chem 283:35236–35246. https://doi.org/10.1074/jbc.M805634200
Otomo T, Otomo C, Tomchick DR et al (2005) Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol Cell 18:273–281. https://doi.org/10.1016/j.molcel.2005.04.002
Rose R, Weyand M, Lammers M et al (2005) Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature 435:513–518. https://doi.org/10.1038/nature03604
Lammers M, Rose R, Scrima A, Wittinghofer A (2005) The regulation of mDia1 by autoinhibition and its release by Rho-GTP. EMBO J 24:4176–4187. https://doi.org/10.1038/sj.emboj.7600879
Kühn S, Geyer M (2014) Formins as effector proteins of Rho GTPases. Small GTPases 5:e983876. https://doi.org/10.4161/sgtp.29513
Labat-de-Hoz L, Alonso MA (2020) The formin INF2 in disease: Progress from 10 years of research. Cell Mol Life Sci 77:4581–4600. https://doi.org/10.1007/s00018-020-03550-7
Brown EJ, Schlondorff JS, Becker DJ et al (2010) Mutations in the formin gene INF2 cause focal segmental glomerulosclerosis. Nat Genet 42:72–76. https://doi.org/10.1038/ng.505
Barua M, Brown EJ, Charoonratana VT et al (2013) Mutations in the INF2 gene account for a significant proportion of familial but not sporadic focal and segmental glomerulosclerosis. Kidney Int 83:316–322. https://doi.org/10.1038/ki.2012.349
Boyer O, Nevo F, Plaisier E et al (2011) INF2 mutations in Charcot-Marie-Tooth disease with glomerulopathy. N Engl J Med 365:2377–2388. https://doi.org/10.1056/NEJMoa1109122
Rossor AM, Polke JM, Houlden H, Reilly MM (2013) Clinical implications of genetic advances in Charcot-Marie-Tooth disease. Nat Rev Neurol 9:562–571. https://doi.org/10.1038/nrneurol.2013.179
Chhabra ES, Higgs HN (2006) INF2 is a WASP homology 2 motif-containing formin that severs actin filaments and accelerates both polymerization and depolymerization. J Biol Chem 281:26754–26767. https://doi.org/10.1074/jbc.M604666200
Madrid R, Aranda JF, Rodríguez-Fraticelli AE et al (2010) The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev Cell 18:814–827. https://doi.org/10.1016/j.devcel.2010.04.001
Shao X, Li Q, Mogilner A et al (2015) Mechanical stimulation induces formin-dependent assembly of a perinuclear actin rim. Proc Natl Acad Sci USA 112:E2595–E2601. https://doi.org/10.1073/pnas.1504837112
Wales P, Schuberth CE, Aufschnaiter R et al (2016) Calcium-mediated actin reset (CaAR) mediates acute cell adaptations. Elife 5:e19850. https://doi.org/10.7554/eLife.19850
Bayraktar S, Nehrig J, Menis E et al (2020) A deregulated stress response underlies distinct INF2-associated disease profiles. J Am Soc Nephrol 31:1296–1313. https://doi.org/10.1681/ASN.2019111174
Rodriguez-Boulan E, Kreitzer G, Musch A (2005) Organization of vesicular trafficking in epithelia. Nat Rev Mol Cell Biol 6:233–247. https://doi.org/10.1038/nrm1593
Rangel L, Bernabe-Rubio M, Fernandez-Barrera J et al (2019) Caveolin-1α regulates primary cilium length by controlling RhoA GTPase activity. Sci Rep 9:1116. https://doi.org/10.1038/s41598-018-38020-5
Isogai T, Innocenti M (2016) New nuclear and perinuclear functions of formins. Biochem Soc Trans 44:1701–1708. https://doi.org/10.1042/BST20160187
Davidson PM, Cadot B (2021) Actin on and around the nucleus. Trends Cell Biol 31:211–223. https://doi.org/10.1016/j.tcb.2020.11.009
Ramabhadran V, Korobova F, Rahme GJ, Higgs HN (2011) Splice variant-specific cellular function of the formin INF2 in maintenance of Golgi architecture. Mol Biol Cell 22:4822–4833. https://doi.org/10.1091/mbc.E11-05-0457
Wang Y, Sherrard A, Zhao B et al (2019) GPCR-induced calcium transients trigger nuclear actin assembly for chromatin dynamics. Nat Commun 10:5271. https://doi.org/10.1038/s41467-019-13322-y
Dürvanger Z, Harmat V (2019) Structural diversity in calmodulin—peptide interactions. Curr Protein Pept Sci 20:1102–1111. https://doi.org/10.2174/1389203720666190925101937
Mruk K, Farley BM, Ritacco AW, Kobertz WR (2014) Calmodulation meta-analysis: predicting calmodulin binding via canonical motif clustering. J Gen Physiol 144:105–114. https://doi.org/10.1085/jgp.201311140
Zhang M, Abrams C, Wang L et al (2012) Structural basis for calmodulin as a dynamic calcium sensor. Structure 20:911–923. https://doi.org/10.1016/j.str.2012.03.019
Chen T-W, Wardill TJ, Sun Y et al (2013) Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499:295–300. https://doi.org/10.1038/nature12354
Bubb MR, Spector I, Beyer BB, Fosen KM (2000) Effects of jasplakinolide on the kinetics of actin polymerization. J Biol Chem 275:5163–5170. https://doi.org/10.1074/jbc.275.7.5163
Chakrabarti R, Ji W-K, Stan RV et al (2018) INF2-mediated actin polymerization at the ER stimulates mitochondrial calcium uptake, inner membrane constriction, and division. J Cell Biol 217:251–268. https://doi.org/10.1083/jcb.201709111
Posern G, Treisman R (2006) Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol 16:588–596. https://doi.org/10.1016/j.tcb.2006.09.008
Olson EN, Nordheim A (2010) Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol 11:353–365. https://doi.org/10.1038/nrm2890
Miralles F, Posern G, Zaromytidou A-I, Treisman R (2003) Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113:329–342. https://doi.org/10.1016/S0092-8674(03)00278-2
Fernandez-Barrera J, Bernabe-Rubio M, Casares-Arias J et al (2018) The actin-MRTF-SRF transcriptional circuit controls tubulin acetylation via α-TAT1 gene expression. J Cell Biol 217:929–944. https://doi.org/10.1083/jcb.201702157
Chhabra ES, Ramabhadran V, Gerber SA, Higgs HN (2009) INF2 is an endoplasmic reticulum-associated formin protein. J Cell Sci 122:1430–1440. https://doi.org/10.1242/jcs.040691
Mu A, Fung TS, Kettenbach AN et al (2019) A complex containing lysine-acetylated actin inhibits the formin INF2. Nat Cell Biol 21:592–602. https://doi.org/10.1038/s41556-019-0307-4
Mu A, Fung TS, Francomacaro LM et al (2020) Regulation of INF2-mediated actin polymerization through site-specific lysine acetylation of actin itself. Proc Natl Acad Sci USA 117:439–447. https://doi.org/10.1073/pnas.1914072117
Høyer-Hansen M, Jäättelä M (2007) Connecting endoplasmic reticulum stress to autophagy by unfolded protein response and calcium. Cell Death Differ 14:1576–1582. https://doi.org/10.1038/sj.cdd.4402200
Wood W (2012) Wound healing: calcium flashes illuminate early events. Curr Biol 22:R14–R16. https://doi.org/10.1016/j.cub.2011.11.019
Chin D, Means AR (2000) Calmodulin: a prototypical calcium sensor. Trends Cell Biol 10:322–328. https://doi.org/10.1016/S0962-8924(00)01800-6
Crivici A, Ikura M (1995) Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct 24:85–116. https://doi.org/10.1146/annurev.bb.24.060195.000505
Kage F, Vicente-Manzanares M, McEwan BC et al (2022) Myosin II proteins are required for organization of calcium-induced actin networks upstream of mitochondrial division. Mol Biol Cell 33:ar63. https://doi.org/10.1091/mbc.E22-01-0005
Liu Z, Wei Y, Zhang L et al (2019) Induction of store-operated calcium entry (SOCE) suppresses glioblastoma growth by inhibiting the Hippo pathway transcriptional coactivators YAP/TAZ. Oncogene 38:120–139. https://doi.org/10.1038/s41388-018-0425-7
Takeuchi Y, Narumi R, Akiyama R et al (2020) Calcium wave promotes cell extrusion. Curr Biol 30:670-681.e6. https://doi.org/10.1016/j.cub.2019.11.089
Yang C, Svitkina TM (2019) Ultrastructure and dynamics of the actin−myosin II cytoskeleton during mitochondrial fission. Nat Cell Biol 21:603–613. https://doi.org/10.1038/s41556-019-0313-6
Andres-Delgado L, Anton OM, Madrid R et al (2010) Formin INF2 regulates MAL-mediated transport of Lck to the plasma membrane of human T lymphocytes. Blood 116:5919–5929. https://doi.org/10.1182/blood-2010-08-300665
Agromayor M, Carlton JG, Phelan JP et al (2009) Essential role of hIST1 in cytokinesis. Mol Biol Cell 20:1374–1387. https://doi.org/10.1091/mbc.e08-05-0474
Delaglio F, Grzesiek S, GeertenW V et al (1995) NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. https://doi.org/10.1007/BF00197809
Orekhov VYu, Jaravine VA (2011) Analysis of non-uniformly sampled spectra with multi-dimensional decomposition. Prog Nucl Magn Reson Spectrosc 59:271–292. https://doi.org/10.1016/j.pnmrs.2011.02.002
Koradi R, Billeter M, Wüthrich K (1996) MOLMOL: A program for display and analysis of macromolecular structures. J Mol Graph 14:51–55. https://doi.org/10.1016/0263-7855(96)00009-4
Chang A, Abderemane-Ali F, Hura GL et al (2018) A calmodulin C-lobe Ca2+-dependent switch governs Kv7 channel function. Neuron 97:836-852.e6. https://doi.org/10.1016/j.neuron.2018.01.035
Fields GB, Noble RL (2009) Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int J Pept Protein Res 35:161–214. https://doi.org/10.1111/j.1399-3011.1990.tb00939.x
Rich RL, Myszka DG (2000) Survey of the 1999 surface plasmon resonance biosensor literature. J Mol Recognit 13:388–407. https://doi.org/10.1002/1099-1352(200011/12)13:6%3c388::AID-JMR516%3e3.0.CO;2-#
Agamasu C, Ghirlando R, Taylor T et al (2019) KRAS prenylation is required for bivalent binding with calmodulin in a nucleotide-independent manner. Biophys J 116:1049–1063. https://doi.org/10.1016/j.bpj.2019.02.004
Sivashanmugam A, Murray V, Cui C et al (2009) Practical protocols for production of very high yields of recombinant proteins using Escherichia coli. Protein Sci 18:936–948. https://doi.org/10.1002/pro.102
Blommel PG, Fox BG (2007) A combined approach to improving large-scale production of tobacco etch virus protease. Protein Expr Purif 55:53–68. https://doi.org/10.1016/j.pep.2007.04.013
Hwang TL, Shaka AJ (1995) Water suppression that works. Excitation sculpting using arbitrary wave-forms and pulsed-field gradients. J Magn Reson A 112:275–279. https://doi.org/10.1006/jmra.1995.1047
Markley JL, Bax A, Arata Y et al (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB inter-union task group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. Eur J Biochem 256:1–15. https://doi.org/10.1046/j.1432-1327.1998.2560001.x
Lee W, Tonelli M, Markley JL (2015) NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinforma Oxf Engl 31:1325–1327. https://doi.org/10.1093/bioinformatics/btu830
Wishart DS, Bigam CG, Yao J et al (1995) 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6:135–140. https://doi.org/10.1007/BF00211777
Morales P, Jiménez MA (2019) Design and structural characterisation of monomeric water-soluble α-helix and β-hairpin peptides: state-of-the-art. Arch Biochem Biophys 661:149–167. https://doi.org/10.1016/j.abb.2018.11.014
Güntert P (2004) Automated NMR structure calculation with CYANA. Protein NMR techniques. Humana Press, pp 353–378
Shen Y, Bax A (2013) Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR 56:227–241. https://doi.org/10.1007/s10858-013-9741-y
Schumann FH, Riepl H, Maurer T et al (2007) Combined chemical shift changes and amino acid specific chemical shift mapping of protein–protein interactions. J Biomol NMR 39:275–289. https://doi.org/10.1007/s10858-007-9197-z
Dominguez C, Boelens R, Bonvin AMJJ (2003) HADDOCK: A protein−protein docking approach based on biochemical or biophysical information. J Am Chem Soc 125:1731–1737. https://doi.org/10.1021/ja026939x
van Zundert GCP, Rodrigues JPGLM, Trellet M et al (2016) The HADDOCK2.2 Web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol 428:720–725. https://doi.org/10.1016/j.jmb.2015.09.014
Acknowledgements
The expert technical advice of the Optical and Confocal Microscopy Unit of the Centro de Biología Molecular Severo Ochoa (CSIC-UAM) is gratefully acknowledged. We thank P Mason for revising the English language of the manuscript, and S Aguilar for her work in the initial stages of this study. NMR experiments were performed in the Manuel Rico NMR laboratory (LMR) of the Spanish National Research Council (CSIC), a node of the Spanish Large-Scale National Facility (ICTS R-LRB). We thank M Lombardía of the Proteomics Facility of the Centro Nacional de Biotecnología (CNB, CSIC) for peptide synthesis.
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. This work was supported by grants PID2021-123179NB-I00 from the Spanish Ministerio de Ciencia e Innovación/Agencia Estatal de Investigación (MCIN/AEI/https://doi.org/10.13039/501100011033) and FEDER Una manera de hacer Europa, and 2021AEP011 from CSIC to MAA, and a grant (PID2020-112821 GB-I00) from MCIN/AEI/https://doi.org/10.13039/501100011033 to MAJ. Research in IC’s and MAJ’s laboratories was supported by a grant (B2017/BMD-3817) from the Comunidad de Madrid, Spain. LK’s laboratory was supported by an intramural grant from CSIC (201720E09) and a grant (PID2019-105404RB-I00) from MCIN/AEI/https://doi.org/10.13039/501100011033. Contracts to AR-R (BES-2016–079136) and LL-d-H (FPU16/00935) from the MCIN and Ministerio de Universidades, respectively, are also acknowledged.
Author information
Authors and Affiliations
Contributions
LL-de-H performed the experiments in cells, made some of the constructs and performed most of the pull-down assays; LC expressed and purified the protein required for NMR experiments and performed the NMR analysis; AR-R, JC-A and LF-M prepared some of the constructs and collaborated in the pull-down experiments; DP-U acquired and processed the NMR data; MTM performed the surface plasmon resonance experiments; LK supervised the surface plasmon resonance experiments and, together with MTM, wrote the corresponding part of the manuscript; MAJ supervised the NMR analyses and wrote the part of the manuscript concerning the NMR analyses; IC, supervised the work and helped writing the manuscript; MAA designed and supervised the work, and wrote the manuscript, with input from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Video 1. Effect of the expression of normal INF2 or INF2 proteins with CaMBS mutations on actin dynamics in INF2 KO cells. Control GCaMP6S cells or INF2 KO GCaMP6S cells expressing or not mCherry fusions of normal INF2 or the INF2 W11L14L18A mutant were stained with SiR-Actin and treated with A23187 (0’). The signal corresponding to GCaMP6S and SiR-Actin is shown
Video 2. Effect of the expression of normal INF2 or the W11L14L18A INF2 mutant on MRTF-A distribution. INF2 KO GFP-MRTF-A cells expressing mCherry fusions of normal INF2 or the W11L14L18A INF2 mutant were treated with A23187 (0’). The signal corresponding to GFP-MRTF-A is shown
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Labat-de-Hoz, L., Comas, L., Rubio-Ramos, A. et al. Structure and function of the N-terminal extension of the formin INF2. Cell. Mol. Life Sci. 79, 571 (2022). https://doi.org/10.1007/s00018-022-04581-y
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04581-y