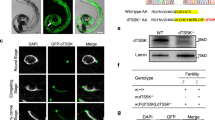
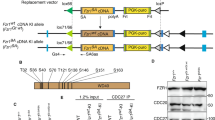
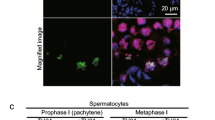
Abstract
Meiosis, a highly conserved process in organisms from fungi to mammals, is subjected to protein phosphorylation regulation. Due to the low abundance of phosphorylation, there is a lack of systemic characterization of phosphorylation regulation of meiosis in mammals. Using the phosphoproteomic approach, we profiled large-scale phosphoproteome of purified primary spermatocytes undergoing meiosis I, and identified 14,660 phosphorylation sites in 4419 phosphoproteins. Kinase-substrate phosphorylation network analysis followed by in vitro meiosis study showed that CDK9 was essential for meiosis progression to metaphase I and had enriched substrate phosphorylation sites in proteins involved in meiotic cell cycle. In addition, histones and epigenetic factors were found to be widely phosphorylated. Among those, HASPIN was found to be essential for male fertility. Haspin knockout led to misalignment of chromosomes, apoptosis of metaphase spermatocytes and a decreased number of sperm by deregulation of H3T3ph, chromosomal passenger complex (CPC) and spindle assembly checkpoint (SAC). The complicated protein phosphorylation and its important regulatory functions in meiosis indicated that in-depth studies of phosphorylation-mediated signaling could help us elucidate the mechanisms of meiosis.






Similar content being viewed by others
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX [66] partner repository with the dataset identifier PXD032089 (password: rp45).
Abbreviations
- CDK:
-
Cyclin-dependent kinase
- HASPIN:
-
Haploid germ cell-specific nuclear protein
- H3T3:
-
Histone H3 threonine 3
- CPC:
-
Chromosomal passenger complex
- SAC:
-
Spindle assembly checkpoint
- PP2A:
-
Serine-threonine protein phosphatase 2A
- H2AX:
-
H2A Histone family, member X
- SYCP3:
-
Synaptonemal complex protein 3
- Ti4+-IMAC:
-
Titanium (IV) ion-immobilized metal affinity chromatography
- LC–MS:
-
Liquid chromatography-mass spectrometry
- REC8:
-
Meiotic recombination protein REC8 homolog
- STAG3:
-
Cohesin subunit SA-3
- SMC3:
-
Structural maintenance of chromosomes protein 3
- SMC1:
-
Structural maintenance of chromosomes protein 1
- KSPN:
-
Kinase-substrate phosphorylation network
- iGPS:
-
GPS algorithm with the interaction filter
- SRPK1:
-
Serine/arginine-rich protein-specific kinase 1
- OA:
-
Okadaic acid
- H3S10:
-
Histone H3 serine 10
- H1FNT:
-
Testis-specific H1 histone
- H1T:
-
Testicular H1 histone
- HILS1:
-
Putative spermatid-specific linker histone H1-like protein
- PcG:
-
Polycomb group
- TUNEL:
-
TdT-mediated dUTP nick end labeling
- Aurora B:
-
Serine/threonine-protein kinase aurora B
- INCENP:
-
Inner centromere protein
- BUBR1:
-
Bub1-related kinase
- CENP-E:
-
Centromere-associated protein E
- MCAK:
-
Mitotic centromere-associated kinesin
- PTM:
-
Post-translational modification
- DOT1L:
-
DOT1 like histone lysine methyltransferase
- EZH1:
-
Enhancer of zeste homolog 1
- VRK1:
-
Serine/threonine-protein kinase VRK1
- KAT6B:
-
Histone acetyltransferase KAT6B
- KDM7C:
-
Histone-lysine demethylase 7C
- ARID5B:
-
AT-rich interactive domain-containing protein 5B
References
Marston AL, Amon A (2004) Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol 5:983–997
Zickler D, Kleckner N (2015) Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 7:a016626
Liu D, Liao C, Wolgemuth DJ (2000) A role for cyclin A1 in the activation of MPF and G2-M transition during meiosis of male germ cells in mice. Dev Biol 224:388–400
Guo FZ, Zhang LS, Wei JL, Ren LH, Zhang J, Jing L et al (2016) Endosulfan inhibiting the meiosis process via depressing expressions of regulatory factors and causing cell cycle arrest in spermatogenic cells. Environ Sci Pollut Res Int 23:20506–20516
Du M, Yuan L, Zhang Z, Zhang C, Zhu M, Zhang Z et al (2021) PPP2R1B is modulated by ubiquitination and is essential for spermatogenesis. FASEB J 35:e21564
Chauhan S, Diril MK, Lee JH, Bisteau X, Manoharan V, Adhikari D et al (2016) Cdk2 catalytic activity is essential for meiotic cell division in vivo. Biochem J 473:2783–2798
Fan Y, Cheng Y, Li Y, Chen B, Wang Z, Wei T et al (2020) Phosphoproteomic analysis of neonatal regenerative myocardium revealed important roles of checkpoint kinase 1 via activating mammalian target of rapamycin C1/ribosomal protein S6 kinase b-1 pathway. Circulation 141:1554–1569
Li L, Zhu S, Shu W, Guo Y, Guan Y, Zeng J et al (2020) Characterization of metabolic patterns in mouse oocytes during meiotic maturation. Mol Cell 80:525–540
Qi L, Liu Z, Wang J, Cui Y, Guo Y, Zhou T et al (2014) Systematic analysis of the phosphoproteome and kinase-substrate networks in the mouse testis. Mol Cell Proteomics 13:3626–3638
Wang M, Guo Y, Wang M, Zhou T, Xue Y, Du G et al (2017) The glial cell-derived neurotrophic factor (GDNF)-responsive phosphoprotein landscape identifies raptor phosphorylation required for spermatogonial progenitor cell proliferation. Mol Cell Proteomics 16:982–997
Li Y, Cheng Y, Zhu T, Zhang H, Li W, Guo Y et al (2019) The protein phosphorylation landscape of mouse spermatids during spermiogenesis. Proteomics 19:e1900055
Villen J, Gygi SP (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat Protoc 3:1630–1638
Hornbeck PV, Kornhauser JM, Latham V, Murray B, Nandhikonda V, Nord A et al (2019) 15 years of PhosphoSitePlus(R): integrating post-translationally modified sites, disease variants and isoforms. Nucleic Acids Res 47:D433–D441
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14:1188–1190
Reyes C, Serrurier C, Gauthier T, Gachet Y, Tournier S (2015) Aurora B prevents chromosome arm separation defects by promoting telomere dispersion and disjunction. J Cell Biol 208:713–727
Song C, Ye M, Liu Z, Cheng H, Jiang X, Han G et al (2012) Systematic analysis of protein phosphorylation networks from phosphoproteomic data. Mol Cell Proteomics 11:1070–1083
Wang L, Liu W, Zhao W, Song G, Wang G, Wang X et al (2014) Phosphorylation of CDK2 on threonine 160 influences silencing of sex chromosome during male meiosis. Biol Reprod 90:138
Willette RN, Eybye ME, Olzinski AR, Behm DJ, Aiyar N, Maniscalco K et al (2009) Differential effects of p38 mitogen-activated protein kinase and cyclooxygenase 2 inhibitors in a model of cardiovascular disease. J Pharmacol Exp Ther 330:964–970
Zhang XM, Zhang L, Wang G, Niu W, He Z, Ding L et al (2015) Suppression of mitochondrial fission in experimental cerebral ischemia: the potential neuroprotective target of p38 MAPK inhibition. Neurochem Int 90:1–8
Pfister, KB et al “Heteroaryl compounds as kinase inhibitors.” US, US 2012/0157433 A1
Fukuhara T, Hosoya T, Shimizu S, Sumi K, Oshiro T, Yoshinaka Y et al (2006) Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc Natl Acad Sci USA 103:11329–11333
Di Agostino S, Botti F, Di Carlo A, Sette C, Geremia R (2004) Meiotic progression of isolated mouse spermatocytes under simulated microgravity. Reproduction 128:25–32
Jordan PW, Karppinen J, Handel MA (2012) Polo-like kinase is required for synaptonemal complex disassembly and phosphorylation in mouse spermatocytes. J Cell Sci 125:5061–5072
Jiang H, Gao Q, Zheng W, Yin S, Wang L, Zhong L et al (2018) MOF influences meiotic expansion of H2AX phosphorylation and spermatogenesis in mice. PLoS Genet 14:e1007300
Medvedeva YA, Lennartsson A, Ehsani R, Kulakovskiy IV, Vorontsov IE, Panahandeh P et al (2015) EpiFactors: a comprehensive database of human epigenetic factors and complexes. Database 2015:v67
Guo X, Zhang P, Qi Y, Chen W, Chen X, Zhou Z et al (2011) Proteomic analysis of male 4C germ cell proteins involved in mouse meiosis. Proteomics 11:298–308
Gohbara A, Katagiri K, Sato T, Kubota Y, Kagechika H, Araki Y et al (2010) In vitro murine spermatogenesis in an organ culture system. Biol Reprod 83:261–267
Chen Y, Zheng Y, Gao Y, Lin Z, Yang S, Wang T et al (2018) Single-cell RNA-seq uncovers dynamic processes and critical regulators in mouse spermatogenesis. Cell Res 28:879–896
Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A (1988) The proto-oncogene c-kit encoding a transmembrane tyrosine kinase receptor maps to the mouse W locus. Nature 335:88–89
Hindriksen S, Lens S, Hadders MA (2017) The ins and outs of aurora B inner centromere localization. Front Cell Dev Biol 5:112
Xie J, Wooten M, Tran V, Chen BC, Pozmanter C, Simbolon C et al (2015) Histone H3 threonine phosphorylation regulates asymmetric histone inheritance in the drosophila male germline. Cell 163:920–933
Parra MT, Viera A, Gomez R, Page J, Carmena M, Earnshaw WC et al (2003) Dynamic relocalization of the chromosomal passenger complex proteins inner centromere protein (INCENP) and aurora-B kinase during male mouse meiosis. J Cell Sci 116:961–974
Parra MT, Gomez R, Viera A, Llano E, Pendas AM, Rufas JS et al (2009) Sequential assembly of centromeric proteins in male mouse meiosis. PLoS Genet 5:e1000417
Kimmins S, Crosio C, Kotaja N, Hirayama J, Monaco L, Hoog C et al (2007) Differential functions of the Aurora-B and Aurora-C kinases in mammalian spermatogenesis. Mol Endocrinol 21:726–739
Carmena M, Wheelock M, Funabiki H, Earnshaw WC (2012) The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol 13:789–803
Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L et al (2004) Aurora B regulates MCAK at the mitotic centromere. Dev Cell 6:253–268
Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S et al (2012) A map of the cis-regulatory sequences in the mouse genome. Nature 488:116–120
Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R et al (2003) Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet 35:25–31
Satyanarayana A, Berthet C, Lopez-Molina J, Coppola V, Tessarollo L, Kaldis P (2008) Genetic substitution of Cdk1 by Cdk2 leads to embryonic lethality and loss of meiotic function of Cdk2. Development 135:3389–3400
Oqani RK, Lin T, Lee JE, Choi KM, Shin HY, Jin DI (2016) P-TEFb Kinase activity is essential for global transcription, resumption of meiosis and embryonic genome activation in pig. PLoS ONE 11:e152254
Wang H, Jo YJ, Sun TY, Namgoong S, Cui XS, Oh JS et al (2016) Inhibition of CDK7 bypasses spindle assembly checkpoint via premature cyclin B degradation during oocyte meiosis. Biochim Biophys Acta 1863:2993–3000
Li J, Zhang HY, Wang F, Sun QY, Qian WP (2021) The Cyclin B2/CDK1 complex conservatively inhibits separase activity in oocyte meiosis II. Front Cell Dev Biol 9:648053
Lowndes NF, Toh GW (2005) DNA repair: the importance of phosphorylating histone H2AX. Curr Biol 15:R99–R102
Kim S, Kim NH, Park JE, Hwang JW, Myung N, Hwang KT et al (2020) PRMT6-mediated H3R2me2a guides Aurora B to chromosome arms for proper chromosome segregation. Nat Commun 11:612
Cha TL, Zhou BP, Xia W, Wu Y, Yang CC, Chen CT et al (2005) Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310:306–310
Baba A, Ohtake F, Okuno Y, Yokota K, Okada M, Imai Y et al (2011) PKA-dependent regulation of the histone lysine demethylase complex PHF2-ARID5B. Nat Cell Biol 13:668–675
Dai J, Sultan S, Taylor SS, Higgins JM (2005) The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev 19:472–488
Soupsana K, Karanika E, Kiosse F, Christogianni A, Sfikas Y, Topalis P et al (2021) Distinct roles of haspin in stem cell division and male gametogenesis. Sci Rep 11:19901
Berenguer I, López-Jiménez P, Mena I, et al (2021) Haspin participates in Aurora phosphorylation at centromeres and contributes to chromosome congression in male mouse meiosis. Cold Spring Harb Lab
Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM (2004) Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol 6:232–237
Petersen J, Hagan IM (2003) S. pombe aurora kinase/survivin is required for chromosome condensation and the spindle checkpoint attachment response. Curr Biol 13:590–597
Ritter A, Sanhaji M, Steinhauser K, Roth S, Louwen F, Yuan J (2015) The activity regulation of the mitotic centromere-associated kinesin by Polo-like kinase 1. Oncotarget 6:6641–6655
Zhang Y, Guo R, Cui Y, Zhu Z, Zhang Y, Wu H et al (2017) An essential role for PNLDC1 in piRNA 3′ end trimming and male fertility in mice. Cell Res 27:1392–1396
Bellve AR (1993) Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol 225:84–113
Tyanova S, Temu T, Cox J (2016) The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 11:2301–2319
UniProt Consortium (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506–D515
Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z et al (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2:100141
Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S et al (2012) A travel guide to cytoscape plugins. Nat Methods 9:1069–1076
Thissen D, Steinberg L, Kuang D (2002) Quick and easy implementation of the Benjamini-Hochberg procedure for controlling the false positive rate in multiple comparisons. J. Educ Behav Stat 27:77–83
Bult CJ, Blake JA, Smith CL, Kadin JA, Richardson JE, Anagnostopoulos A et al (2019) Mouse genome database (MGD) 2019. Nucleic Acids Res 47:D801–D806
Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S et al (2021) The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49:D605–D612
Newby LK, Marber MS, Melloni C, Sarov-Blat L, Aberle LH, Aylward PE et al (2014) Losmapimod, a novel p38 mitogen-activated protein kinase inhibitor, in non-ST-segment elevation myocardial infarction: a randomised phase 2 trial. Lancet 384:1187–1195
Krulikas LJ, McDonald IM, Lee B, Okumu DO, East MP, Gilbert T et al (2018) Application of integrated drug screening/kinome analysis to identify inhibitors of gemcitabine-resistant pancreatic cancer cell growth. SLAS Discov 23:850–861
Zhu F, Li W, Zhou X, Chen X, Zheng M, Cui Y et al (2021) PRSS55 plays an important role in the structural differentiation and energy metabolism of sperm and is required for male fertility in mice. J Cell Mol Med 25:2040–2051
Qi M, Sun H, Guo Y, Zhou Y, Gu X, Jin J et al (2022) m(6) A reader protein YTHDF2 regulates spermatogenesis by timely clearance of phase-specific transcripts. Cell Prolif 55:e13164
Ma J, Chen T, Wu S, Yang C, Bai M, Shu K et al (2019) iProX: an integrated proteome resource. Nucleic Acids Res 47:D1211–D1217
Funding
This study was supported by the National Key R&D Program of China (2021YFC2700200), the Chinese National Natural Science Foundation (Grants Nos. 32071133, 82001611, 81971439), and the Fok Ying Tung Education Foundation (Grant No. 161037).
Author information
Authors and Affiliations
Contributions
XG, CS and YL conceived and designed the study. HL, HC, XZ, YQ, YW and LY performed most of the experiments. XZ and YG performed mass spectrometry experiments. BW conducted all of the mass spectrometry data analysis. YC constructed mouse models. JR, YZ, TZ, YC and HZ, analyzed the data. YL, CS and XG supervised the project. HL, YL, CS and XG wrote the manuscript, which was reviewed by all authors.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval
Animal experiments were performed in compliance with ethical regulations of Institutional Animal Care and Use Committee (IACUC) of Nanjing Medical University (No: IACUC-1707017).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Li, H., Chen, H., Zhang, X. et al. Global phosphoproteomic analysis identified key kinases regulating male meiosis in mouse. Cell. Mol. Life Sci. 79, 467 (2022). https://doi.org/10.1007/s00018-022-04507-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04507-8