Abstract
Since decades it has been known that non-protein-coding RNAs have important cellular functions. Deep sequencing recently facilitated the discovery of thousands of novel transcripts, now classified as long noncoding RNAs (lncRNAs), in many vertebrate and invertebrate species. LncRNAs are involved in a wide range of cellular mechanisms, from almost all aspects of gene expression to protein translation and stability. Recent findings implicate lncRNAs as key players of cellular differentiation, cell lineage choice, organogenesis and tissue homeostasis. Moreover, lncRNAs are involved in pathological conditions such as cancer and cardiovascular disease, and therefore provide novel biomarkers and pharmaceutical targets. Here we discuss examples illustrating the versatility of lncRNAs in gene control, development and differentiation, as well as in human disease.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
It has long been known that several classes of non-protein-coding RNA molecules exert important cellular functions. For instance, ribosomal RNAs (rRNAs) are essential elements of the translation machinery and small nuclear RNAs (snRNAs) are required for splicing of nascent RNA transcripts. Also, various classes of small (around 20–30 nucleotides) noncoding RNAs such as micro (mi)RNAs, small inhibitory (si)RNAs or PIWI interacting (pi)RNAs are well known as gene silencers. With the recent advent of massive parallel sequencing techniques, however, it has been observed that a tremendously high portion, approximately 70 %, of the genome is transcribed in various contexts and cell types [1, 2]. A large proportion of these newly detected RNA transcripts are structurally indistinguishable from protein-coding and processed messenger RNAs (mRNAs). They tend to be expressed at a very low level and have little to no protein-coding potential. This subclass of noncoding transcripts of variable length and function is collectively referred to as long noncoding RNAs (lncRNAs).
A plethora of biological tissues, organs, pathological samples and cultured cells have been analyzed for noncoding RNA expression, and it is clear that these molecules are omnipresent. Apparently, defining noncoding RNA function has proven more challenging than detecting them, as the number of reports showing comprehensive functional data is far smaller than those describing their identification in various contexts. The flexibility of RNA transcripts and their ability to fold into complex 3D-conformations enables them to form specific interactions with proteins. They can interact with RNA or DNA molecules via base pairing, even with double-stranded DNA, and form networks with DNA, protein complexes and RNA molecules, illustrating their large potential as an important player with many biological functions. In this review, we will discuss mechanisms of lncRNA functions with a focus on their role in development and disease (Table 1).
Molecular and genetic structure of LncRNAs
Like mRNAs, lncRNAs are transcribed by Polymerase II, mostly 5′-capped, polyadenylated and spliced, though on average they contain a lower number of exons than mRNAs and their expression level assessed across different tissues is lower [3, 4]. There are various algorithms calculating the coding probability based on the length of a potential open reading frame (ORF), the similarity of such an ORF to known protein-coding genes, frequency of in-frame nucleotide hexamers or other empirical sequence features [5]. In general, RNA transcripts containing short (<100 nt) non-conserved ORFs, which have no homology to known peptide sequences and do not match to peptides identified in mass spectrometry screens are considered noncoding [6]. Interestingly, the majority of lncRNAs are associated with ribosomes [7, 8], though they do not show the characteristic release of ribosomes [9] or the typical 3-nucleotide phasing corresponding to codons of an ORF [10, 11]. However, in rare instances, functional oligopeptides have been found to be translated from putative lncRNAs [12–14].
Deep-sequencing experiments revealed many examples of genes producing both protein-coding and noncoding transcripts by alternative splicing. However, so far, only very few reports demonstrate a functional role for both the noncoding RNA(s) and the protein(s) encoded by transcripts derived from the same gene [15, 16]. It is tempting to speculate that such dual usage of transcripts is more frequent than anticipated.
RNA molecules have the potential to form highly structured macromolecules by folding into double-stranded stems, single-stranded loops and bulges, which again can fold further into three-dimensional structures, allowing for the potential formation of complex shapes. So far, the structure of only a few RNAs has been experimentally determined using a combination of chemical assays, and by determining the accessibility of base-paired or single-stranded RNA by various RNases [17, 18]. However, these methods still have the limitation that they can only reveal the secondary, but not the tertiary (3D) structure. In addition, computational predictions are only beginning to provide reliable results, but program learning from experimental data might improve the predictions and more closely mirror experimental observations. This process is accelerated by recently developed techniques combined with high-throughput sequencing, such as SHAPE-MaP and icSHAPE [19, 20].
The similarity between mRNA-encoding and lncRNA genes is furthermore reflected by the chromatin signatures at the genomic regions from where they are transcribed. Their transcriptional start site is, in most cases, marked by the H3K4me3 histone modification and the transcribed region by the H3K36me3 mark, no matter if the lncRNA originates from its own promoter or from an enhancer. However, these histone marks are less prominent than observed at mRNA coding genes, whereas H3K4me1, a characteristic mark of enhancers, tends to be more prominent at genomic regions encoding lncRNAs [2, 21–23]. As expected for transcribed regions, lncRNA promoters correspond with DNaseI hypersensitive sites, which indicates accessible chromatin [24, 25].
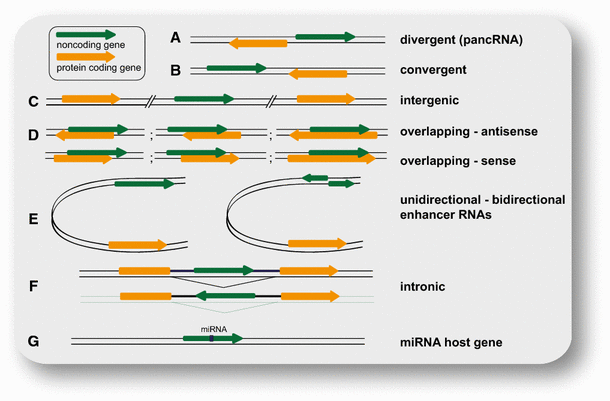
So far, the most commonly used categorization of the highly heterogeneous class of lncRNAs is their position in the genome relative to protein-coding genes (Fig. 1). LncRNAs can be intergenic (lincRNAs) or divergently transcribed from the same promoter as a protein-coding gene (pancRNAs). LincRNAs expressed from a promoter (with high H3K4me3 to H3K4me1 ratio) are classified as promoter-associated lncRNAs (plncRNAs). However, lncRNAs can also be transcribed from enhancers and are then termed eRNAs. They are mostly transcribed in both directions, in contrast to enhancer-associated lncRNAs (elncRNAs), which are unidirectionally transcribed from a promoter with low H3K4me3 to H3K4me1 ratio [23]. Transcription of lncRNAs can also originate from within introns, or overlap with other transcripts in sense or antisense orientation [3]. Some lncRNAs are generated by backsplicing from introns of mRNAs or other lncRNAs and are thus circular (circRNAs) [26, 27].
Classification of lncRNAs according to their position relative to neighboring gene(s). a Divergently transcribed lncRNA originating from the same promoter region as the adjacent (usually protein coding) gene, but from the opposite strand; b convergently transcribed genes encoded on opposite strands and facing each other; c intergenic (or intervening) lncRNA (or lincRNA) located distant from other genes (usually >10 kb); d examples for various cases of lncRNAs overlapping with other genes on the same or the opposite strand; e enhancer RNAs expressed as uni- or bidirectional transcripts; f LncRNA transcribed from an intron of another gene; g lncRNA hosting a miRNA. Noncoding genes are shown in green, protein-coding genes in orange
Genome-wide sequencing of RNA species isolated from cytosolic or nuclear fractions of cells has shown that the majority of lncRNAs tend to be localized in the nucleus or are associated with chromatin, while a considerable fraction localizes to the cytoplasm [3, 28, 29]. The subcellular localization is a good indication of the putative function of a lncRNA, since in contrast to protein-coding mRNAs, lncRNAs can already function while transcription is occurring. In fact, many nuclear and, in particular, chromatin-retained lncRNAs co-regulate transcription and/or chromatin structure at or close to their site of transcription, i.e. in cis.
Conservation and evolutionary aspects of LncRNA
The genomic sequences of lncRNAs are, in general, less conserved than exons, but more than introns of protein-coding genes, pointing towards a rapid evolution with moderate constraints [21, 30]. In contrast to the actual sequence, splice sites of lncRNAs have been found to be more stable during evolution [31, 32]. Despite poor RNA sequence conservation, lncRNAs have frequently been identified across species in syntenic genomic regions [33, 34]. It is therefore quite likely that evolutionary conservation of lncRNAs is embodied by a conserved 3D structure, although this is difficult to assess with current methods. A further layer of conservation is functional conservation between lncRNAs playing equivalent roles in particular biological settings [35].
A different explanation for low conservation of lncRNAs is the increasing number of such transcripts in increasingly complex species. This finding, in combination with the observed highly tissue-specific expression of many lncRNAs, suggests that lncRNAs might be key molecules promoting species-specific features and organ complexity [36–38]. Thus, lncRNAs very likely have played important roles in the evolution of complex organisms.
LncRNAs regulate gene and genome activity
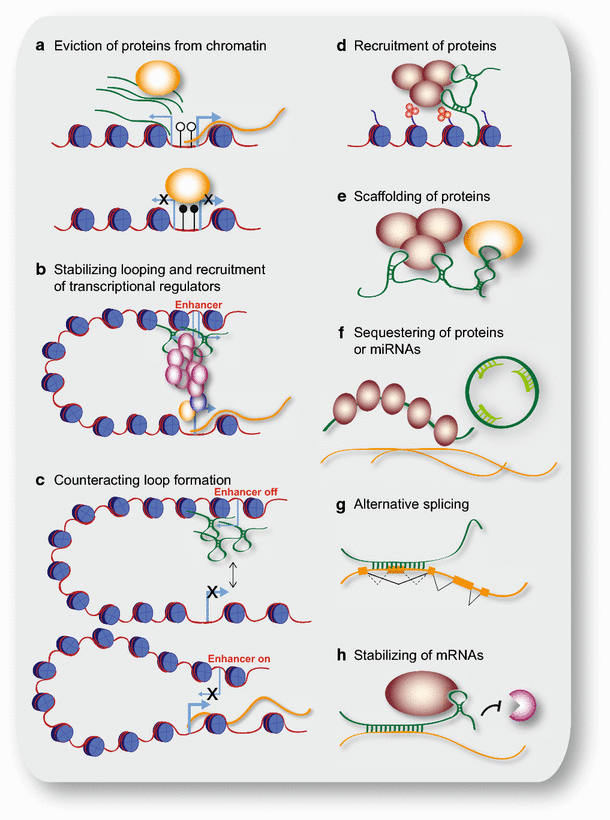
The best-described function for lncRNAs in the nucleus is their role in regulating gene and genome activity on various levels (Fig. 2a–e). Many possible mechanisms by which lncRNAs influence chromatin modifications and chromatin structure, and thereby influence transcription or other chromatin-related functions by epigenetic mechanisms have been investigated. In the following section we will address different levels of gene and genome regulation separately.
Schematic representation of cellular mechanisms involving lncRNAs. a LncRNA transcripts evicting proteins from chromatin; here, pancRNAs prevent DNMT from methylating DNA in their promoter region, thereby ensuring mRNA transcription. b LncRNAs recruiting the Mediator complex to an enhancer region, stabilizing loop formation and transcription of the associated gene. c LncRNAs transcribed from an enhancer region interfering with enhancer-promoter contact, thereby inhibiting transcription of the protein-coding gene. d LncRNA recruiting proteins, such as chromatin-modifying complexes to specific target sites in the genome, e.g. via DNA-RNA triplex formation. e LncRNA acting as scaffold linking different proteins required for concerted action. f LncRNA binding and sequestering proteins to prevent or attenuate their action, e.g. binding to mRNAs (left); circRNA sequestering miRNAs to prevent their binding to mRNAs (right). g Example of a lncRNA changing the splicing pattern by binding to a primary RNA transcript. h LncRNA stabilizing a mRNA by recruiting proteins such as STAU1, thereby preventing degradation
Histone modifications regulated by LncRNAs
Prominent examples of histone-modifying complexes interacting with lncRNAs are the two polycomb repressive complexes, PRC1 and, in particular PRC2, which mediates methylation of lysine 27 on histone 3 (H3K27me), a histone mark associated with repressed or poised genetic loci. The first report of an interaction of PRC2 with a noncoding RNA came from studies on X-chromosome inactivation in mammals, which involves X-inactive specific transcript (Xist), a lncRNA that is highly expressed from the inactive X-chromosomes in females (Xi) and recruits PRC2 to the Xi to silence gene expression [39–41].
Another prominent example is Hotair, a conserved lncRNA that is transcribed from within the HoxC gene cluster. Hotair was shown to play a repressive role at the HoxD locus by interacting with PRC2. In addition, Hotair also interacts with the histone H3K4me1/2 demethylase LSD1 (KDM1), which removes a histone mark of active chromatin and thus reinforces the establishment of a repressive chromatin environment on target loci [42] (Fig. 2d, e). Targeted inactivation of Hotair in mice leads to de-repression of imprinted genes and of HoxD genes, which lose H3K27-methylation and gain H3K4-methylation marks, as expected from loss of the enzymatic activity of PRC2 and LSD1 at these loci. Furthermore, Hotair knockout mice show skeletal homeotic transformation phenotypes, which are typical for mutations affecting Polycomb mediated repression [43]. Interestingly, mice lacking most of the HoxC cluster including Hotair do not display these phenotypes, suggesting a complex interplay of multiple genomic regions in regulating HoxC cluster gene expression [44].
Braveheart (Bvht) is a lncRNA that is activated during early cardiac differentiation and acts upstream of MESP1, a basic Helix-loop-helix transcription factor involved in early heart development. Bvht also exerts its function via an interaction with PRC2 [45]. The lncRNA Fetal-lethal developmental regulatory RNA (Fendrr; or Foxf1 adjacent noncoding developmental regulatory RNA) is likewise involved in cardiac development and heart function and interacts with PRC2. Fendrr also interacts with WDR5, which is well known for its presence in the MLL complexes that mediate H3K4 methylation, a mark that is thought to oppose H3K27me. Fendrr might play a role at poised genes, or is involved in tuning the balance of active and repressive marks at its target gene promoters, thus adjusting the correct expression level of its targets [46, 47]. These examples are only a small selection and many more lncRNAs seem to function in concert with the PRC2 complex; for more examples, see [48–52].
An intriguing mechanism as to how lncRNAs might recruit their protein interaction partners to specific genomic loci is DNA-RNA triplex formation [53], which has been proposed for several lncRNAs forming a complex with PRC2 [46, 54, 55] (Fig. 2d). In this manner, lncRNAs might direct chromatin or transcriptional modulators to specific genomic sites. This could explain, to some extent, the site-specific action of chromatin-modifying complexes, which do not themselves bind to DNA in a sequence specific manner [38, 56].
An example of a lncRNA interacting with the PRC1 complex is Focally amplified lncRNA on Chromosome 1 (FAL1). The interaction of FAL1 with BMI1, an essential subunit of PRC1, regulates its protein stability [57]. The knockdown of FAL1 in an ovarian cancer cell line led to gene expression changes, slower cell cycle progression and induction of senescence, similar to the effects of the BMI1 knockdown. This latter phenotype could be partially rescued by knockdown of the senescence-promoting factor CDKN1A (p21). Additional ChIP data suggested that BMI1 binds to the promoter of CDKN1A and, together with FAL1, suppresses the expression of this target and of many other genes.
Whereas FAL1 acts in trans on PRC1, ANRIL, a lncRNA transcribed from the INK4B-ARF-INK4A tumor suppressor locus, acts in cis. The ANRIL transcript was discovered in a family with inherited melanoma-neural system tumors and assumed to play a role in tumorigenesis [58]. ANRIL recruits CBX7 (a PRC1 component) via a POLII-dependent mechanism to its locus in order to repress the neighboring INK4B-ARF-INK4A genes, antagonize cellular senescence and indirectly promote cell cycle activity [58, 59].
The repressive H3K27-methylation, catalyzed by EZH2 in the PRC2 complex, is opposed by MLL complexes, which mediate methylation of H3K4me2/3, a mark associated with loci that are actively transcribed or primed for activation. WDR5 is an integral part of all MLL complexes, but also interacts with several other protein complexes [60]. WDR5 has been found to interact with more than 200 lncRNAs in mouse embryonic stem cells (mESCs) [61]. These interactions are important for WDR5 binding to chromatin since a point mutant that cannot bind RNA also fails to stably associate with chromatin. More specifically, two lncRNAs, HOTTIP and NeST have been described to recruit WDR5 to their neighboring genes and thus enhance their transcription [62, 63].
LncRNAs modulate DNA methylation
Whereas many lncRNAs associating with Polycomb complexes act by promoting PRC occupancy at genomic target sites, lncRNAs described in the context of DNA methylation have mostly been found to oppose this epigenetic mark. Frequently, transcription of noncoding RNAs, which interact with DNMTs, keeps a locus free of DNA methylation [54, 64–66] (Fig. 2a). Initially, it was observed that knockdown of extracoding CEBPA (ecCEBPA), a transcript starting upstream of the CEBPA gene, leads to down-regulation of CEBPA and increased DNA methylation of the locus. A subsequent global analysis of all lncRNAs bound to DNMT1 [64] revealed that loci whose RNA products interact with DNMT1 show lower levels of DNA methylation than other loci. A similar observation was made in oocytes and two-cell stage embryos [66], in which the down-regulation of several divergently transcribed promoter-associated ncRNA (pancRNA) led to lower expression of the adjacent protein-coding gene and higher DNA methylation. In the case of one particular gene, IL17b, the pancRNA knockdown resulted in cell death of the developing blastocyst. Supplementing the embryos with IL17b protein rescued the phenotype.
In the case of Tcf21 antisense RNA reducing DNA methylation (TARID), keeping the protein-coding gene (Tcf21) on the opposite DNA strand free of DNA methylation required GADD45A and the TET proteins [65] pointing towards a mechanism involving active DNA demethylation.
LncRNAs regulate chromatin remodeling
In addition to interaction with enzymatic complexes, which covalently modify chromatin, lncRNAs have also been shown to control chromatin remodeling complexes that can alter the nucleosome spacing in an energy dependent manner. In human prostate cancer, gene expression analyses showed that SWI/SNF and the lncRNA SChLAP1 have opposing roles. SChlAP1 interacts with the SNF5 subunit of the chromatin remodeling complex SWI/SNF, and globally inhibits binding of SWI/SNF to chromatin, subsequently leading to genome-wide de-repression of gene activity [67]. Similarly, the lncRNA Myheart (Mhrt), a lncRNA transcribed divergently to Myh6 and overlapping with Myh7 in antisense direction, binds to BRG1, the ATPase subunit of the SWI/SNF complex, and excludes it from the Myh6/7 locus, thus preventing chromatin remodeling [68]. BRG1, in turn, down-regulates Mhrt. Cardiac stress leads to upregulation of BRG1 to a level allowing it to overcome the repulsion of the SWI/SNF complex from chromatin by Mhrt and to bind to the Myh6 locus.
The Evf2 lncRNA, transcribed from the genomic region between Dlx5 and Dlx6, promotes SWI/SNF binding to the enhancers of these genes. However, Evf2 inhibits the remodeling activity of SWI/SNF and thus interferes with upregulation of Dlx5/Dlx6. Accordingly, in Evf2 mutant mice, Dlx6 and, to a lesser extent, Dlx5 are upregulated [69, 70]. In a more recent study, it was observed that the binding of Evf2 to BRG1 can be out-competed by other RNAs of similar length and results in reduced remodeling activity, suggesting that the binding of lncRNAs to the SWI/SNF complex is promiscuous. Xist, on the other hand, binds to components of the SWI/SNF complex, but excludes the complex from the Xi, rather than recruiting it [71]. The SWI/SNF complex can also be recruited by lncRNAs and mediate gene activation, as has been found in hepatocellular carcinoma cancer stem cells. A lncRNA, lncTCF7 or WNT signaling pathway activating non-coding RNA (WSPAR), transcribed 200 kb upstream of TCF7, can recruit SWI/SNF to the TCF7 promoter and thus activate TCF7 expression and WNT signaling [72].
Is the observed binding of numerous lncRNAs to chromatin modifiers specific or promiscuous?
The specificity of lncRNA interactions with chromatin-modifying complexes has been questioned. For instance, a large number of lncRNAs have been shown to co-purify with PRC2, possibly suggesting non-specific binding [50, 52, 73]. Similarly, 15 % of all lncRNAs present in a particular cellular context were shown to be bound by DNMT1 [64, 74]. Likewise, WDR5 binds to more than a thousand RNAs in mESCs, of which 20 % are lncRNAs [61]. This issue of promiscuity was assessed exemplarily for PRC2 in a systematic biochemical study [75], where the authors demonstrate that PRC2 binds RNA in a promiscuous manner, but has remarkably higher affinity for specific lncRNAs than for e.g. bacterial mRNA. Furthermore, they provide evidence that the binding strength correlates with the length of the RNA. Though these studies were performed in vitro at non-physiological conditions, they suggest that the chromatin-modifying complexes can sufficiently differentiate between specific and non-specific interactors. Moreover, since most lncRNA transcripts are expressed at few copies per cell, it is likely that the number of PCR2 molecules per cell is in excess to the number of lncRNA transcripts, which contain a high affinity site for PRC2. Overall, it is not unexpected that a chromatin-modifying complex acting at numerous genomic loci binds many different individual lncRNAs, which stabilize it or recruit it to distinct genomic loci.
Interactions of lncRNAs with transcription factors
Besides chromatin modifiers, transcription factors have also been found to interact with lncRNAs. The lncRNA definitive endoderm-associated lncRNA1 (DEANR1) (also known as ALIEN), for instance, is transcribed downstream of FOXA2 and is highly induced during differentiation of ESCs towards definitive endoderm [76, 77]. Knockdown of DEANR1 inhibits endodermal differentiation due to the reduction of FOXA2 expression. This interpretation is strongly supported by the fact that endoderm differentiation is rescued in DEANR1 knockdown cells upon forced FOXA2 expression. Mechanistically, DEANR1 was proposed to interact with SMAD2/3 and to recruit it to the FOXA2 promoter.
The lncRNA rhabdomyosarcoma associated transcript (RMST) is required for neuronal differentiation by mediating binding of the transcription factor SOX2 to approximately half of its binding sites [78]. Certain lncRNAs have been linked in different ways to transcription factors of the NF-κB pathway. Lethe (after the ‘river of forgetfulness’ in Greek mythology) interacts with RelA and prevents it from binding to DNA [79]. p50-associated cyclooxygenase-2 (COX-2) extragenic RNA (PACER), on the other hand, sequesters p50, which forms homodimers and is repressive at high concentration, thereby lowering its concentration and allowing it to form activating heterodimers with p65 [80]. Another lncRNA, lnc dendritic cells (lnc-DC) binds to the transcription factor STAT3 in the cytoplasm and prevents its dephosphorylation by SHP1, thereby activating STAT3 and thus dendritic cell differentiation [81].
An elaborate mechanism linking transcription factor binding and chromatin modifications has been found for breast cancer anti-estrogen resistance 4 (BCAR4). This lncRNA binds to the SMAD nuclear interacting protein 1 (SNIP1) and a phosphatase (PNUTS) involved in RNA polymerase II regulation. In response to cytokine stimulation, BCAR4 lifts the inhibitory effect of SNIP1 on p300, a histone acetyl transferase and transcriptional activator. Acetylated histones are necessary for BCAR4 mediated recruitment of PNUTS, which in turn leads to active polymerase II at GLI2 controlled genes [82].
Also CTCF, a protein with the dual function of a transcriptional regulator and a boundary factor, has been shown to interact with numerous noncoding RNAs [83]. An example is the interplay between CTCF and the lncRNAs Jpx, Tsix and Xite, which is important for X-chromosome inactivation [83, 84]. Prior to inactivation, Tsix and Xite recruit CTCF in cis to the X-inactivation center promoting homologous X-chromosome pairing at the Xic [83]. During X-chromosome inactivation, Jpx evicts CTCF in trans from the Xist locus and thereby prevents promoter blockage by CTCF [83]. These examples illustrate the dynamic interaction between lncRNAs and transcription factors in regulating chromatin-mediated cellular functions.
Regulation of genome organization by looping of enhancers
Previously, transcripts found at enhancers had been interpreted as byproducts of transcription from promoters contacted by enhancers. Now, it is well established that RNA transcripts produced at active enhancers are functionally important [85–87]. An example of enhancer-linked transcripts, already identified two decades ago, has been described for the locus control region (LCR) of the β-globin cluster [88, 89]. To date, numerous studies have assigned functions to enhancer-associated RNAs [90] (Fig. 2b). There are different classes of RNAs observed at enhancers. The classical enhancer RNAs (eRNAs) are derived from bidirectional transcription, are unspliced and rather short transcripts [85, 91]. Additionally, there are also unidirectional, often spliced and processed transcripts, which are indistinguishable from canonical lncRNAs, but happen to be transcribed from an enhancer [23, 92]. These transcripts often function in cis. Finally, there are lncRNAs with enhancer-like functions (also called ncRNA-a) [93]. Whether this classification reflects different functional classes of RNAs remains to be investigated.
In general, eRNAs are co-regulated with their neighboring gene(s) [91] and presumed to be important for gene expression, possibly by supporting cohesin binding [94]. However, experimental support for the latter is rather mild. While eRNAs in some studies were shown to stabilize chromosome loops [94, 95], they appeared to be dispensable for promoter-enhancer contact in others, raising doubts about the role of eRNAs in chromosome looping [96, 97].
LncRNAs transcribed from enhancer regions can also have inhibitory effects on their target gene. The promoter deletion of the lncRNA Haunt (also known as linc-Hoxa1), for instance, has been shown to lead to upregulation of several genes of the neighboring HoxA gene cluster. However, larger deletions in the Haunt locus impair HoxA expression [98], demonstrating that the genomic locus can have opposing functions to the RNA product encoded by it. The repressive effect of Haunt was furthermore observed at the single cell level confirming that the expression of HoxA1 and Haunt are anti-correlated [99].
A similar mechanism was found for Playrr (D030025E07Rik), a lncRNA encoded upstream of the homeodomain transcription factor Pitx2 [100]. Pitx2 expression is constrained to the left side of the gut dorsal mesentery, whereas expression of Playrr is detected on the right, but spreads to both sides in Pitx2 deletion mutants. A CRISPR/Cas9 generated mutation resulting in Playrr RNA decay caused upregulation of Pitx2 expression [100]. Both experiments reveal a mutual inhibitory interaction between Pitx2 and Playrr. An enhancer involved in Pitx2 regulation in the gut overlaps the TSS of Playrr. In the cells on the left-hand side, where Pitx2 is expressed, the Pitx2 locus was found to be in closer proximity to the Playrr locus than in cells on the right side and to cause down-regulation of Playrr by an unknown mechanism. In cells on the right, on the other hand, Playrr RNA is thought to cause a separation of the Pitx2 locus from the enhancer at the Playrr locus suggesting that Playrr expression interferes with the looping of the Pitx2 promoter to its enhancer at the Playrr locus and thus with Pitx2 gene activation (Fig. 2c).
Furthermore, the lncRNAs PRNCR1 and PCGEM1 promote enhancer-promoter looping in prostate cancer cells [101]. Other lncRNAs have been suggested to assist chromosome conformation more globally. A particular situation can be observed at the inactive X-chromosome (Xi). The 3D-structure of Xi is dependent on Xist, supposedly because Xist excludes cohesin binding from Xi [71]. Deletion of Xist led to loss of the compact chromosome structure of the Xi and reorganization into chromosome domains as they are found on the active X-chromosome. Recently, another lncRNA, functional intergenic repeating RNA element (Firre), which escapes X-chromosome inactivation was suggested to stabilize the perinucleolar location of the Xi [102]. Additionally, Firre was proposed to mediate focal trans-chromosomal contacts [103]. This finding was questioned by a different study, which reported a dispersed nuclear distribution of Firre and a role in the control of factors involved in RNA processing [104]. These examples show that lncRNAs can play important roles in controlling local intra- and inter-chromosomal genomic structure and interactions.
Regulation at the posttranscriptional level
LncRNAs primarily found in the cytosol are thought to be involved in gene regulation on the posttranscriptional level (Fig. 2e–h). Accordingly, several lncRNAs have been shown to influence splicing patterns of either specific genes or globally by interacting with splicing factors. The lncRNA Pnky (divergent to Pou3f1) binds to the splicing factor PTBP1 and thereby regulates the splicing patterns of a subset of genes involved in neurogenesis [105]. Whether TUNA (megamind), which interacts with PTBP1 as well, functions via the same mechanism remains to be investigated [33, 106]. In addition, myocardial infarction associated transcript (MIAT) has been found to influence splicing in different systems. In embryonic neurogenesis, MIAT is involved in controlling the splicing pattern of Wnt7b [107]. In iPS cells and mouse primary neurons, MIAT binds to splicing factors, and in post-mortem brains of schizophrenia patients, it was found to be down-regulated, suggesting that it might contribute to this pathological phenotype [108]. MALAT1, a highly expressed lncRNA enriched in nuclear speckles, has been suggested to be a general component of the splicing machinery [109]. However, this view has been challenged since the MALAT1 knockout did not show disintegration of splicing speckles [110]. A very specific role in the cytoplasm was found for the lncRNA non-coding RNA activated by DNA damage (NORAD) [111]. NORAD sequesters PUMILIO proteins and thereby promotes genomic stability (Fig. 2f). Ablation of NORAD leads to hyperactivity of PUMILIO causing chromosome instability and aneuploidy due to repression of PUMILIO target mRNAs that function in DNA replication, mitosis and DNA damage repair.
The most recently described class of lncRNAs that came into focus are the circRNAs. Due to their circular structure, they are resistant to degradation by exonucleases and are therefore thought to be highly stable within a cell. The brain specific circRNA CDR1as was shown to act as a sponge for the miRNA let-7 [26, 27] (Fig. 2f). Accordingly, overexpression of CDR1as in zebrafish caused impaired midbrain development, similarly to the effect of a let-7 inhibitor. It was also shown that small polypeptides can be derived from some circRNAs, extending the functional repertoire of these stable RNA molecules [112]. Most likely, circRNAs do not only interact with other transcripts, but also with proteins and thus may function in a wide range of different processes.
Functions of lncRNAs in development and differentiation
In the following section, we will focus on the physiological effects of lncRNAs in the organism, during embryonic development and in in vitro differentiation systems. There are still few known examples of lncRNAs shown to play important roles in vivo, and thus we will revisit some of the lncRNAs already mentioned above.
Maternal transcripts present in oocytes and zygotes contribute to early differentiation processes. In a single-cell sequencing study examining oocytes, zygotes and cells from the first cell divisions of the early mouse embryo, lncRNA transcripts were found at all stages examined [113]. One subclass of lncRNAs, the pancRNAs, was compared between murine two-cell embryos and oocytes [66]. A subset of pancRNAs found to be expressed in two-cell embryos but not in oocytes was found to be important for counteracting DNA methylation at the adjacent promoters thereby ensuring expression of the neighboring gene.
The mechanisms first found to be linked to lncRNAs were imprinting and X-chromosome inactivation. Accordingly, the first lncRNA that was genetically targeted and deleted was H19, an imprinted gene that was already suspected to be a lncRNA 25 years ago [114]. H19 is exclusively expressed from the maternal allele and is inhibitory for the adjacent gene, Insulin-like growth factor 2 (Igf2) [114–116]. An H19 deletion on the maternally inherited chromosome led to an increase in Igf2 expression and thus to increased bodyweight. This phenotype could be rescued by deletion of one Igf2 allele. H19 knockout mice are viable and fertile [116], but have reduced muscle regeneration capacities due to loss of two miRNAs encoded within the H19 transcript [117]. The imprinted Igf2r gene is silenced by the lncRNA Airn, which is transcribed from the opposite strand, overlapping the 5-prime region of Igf2r. Here, silencing is not achieved by the lncRNA transcripts as such, but by transcription through the Igf2r promoter, which interferes with RNA PolII recruitment [118]. Several further lncRNAs with a well-established role in imprinting have been reported. For details we would like to refer to a recent review [119].
Insight into the developmental function of Xist was obtained two decades ago when Xist knockout mice were generated and analyzed [120, 121]. Male mice lacking Xist are unaffected, whereas females die during the first half of gestation. Interestingly, knockout females with a single X-chromosome (XO) lacking Xist are healthy, strongly suggesting that it is not the lack of Xist, but the failure to adjust the X-chromosome gene dosage that causes embryonic lethality.
In a more recent example, the deletion of another imprinted lncRNA gene in the mouse, Gtl2 (Meg3, maternally expressed 3), showed clear cis-acting effects on neighboring genes resulting in perinatal lethality [122].
Many other mouse models in which lncRNAs have been removed by deletion of genomic fragments did not show overt phenotypes, but after detailed analyses, some specific defects have been observed [110, 123–126]. For instance, deletion of NEAT1 resulted in viable and fertile mice, however, knockout females showed about 50 % reduced fertility due to reduced progesterone production and corpus luteum formation [127]. Ovary transplantation or progesterone administration rescued offspring rates, confirming the cause of the phenotype, though the molecular mechanism remains to be determined.
Fendrr is one of few rare examples of lncRNAs demonstrated to play an essential role in organ development and embryo survival. Fendrr, which is divergently transcribed from the Foxf1 promoter, was analyzed by two different strategies [47, 123]. In one study, the insertion of a transcriptional stop cassette replacing the first exon of Fendrr leaving most of the genomic context undisturbed resulted in impaired heart function, omphalocele (defects in body wall development), and embryonic death [47]. Fendrr interacts with chromatin-modifying complexes and with DNA, possibly through triplex formation, and thereby changes the chromatin landscape of specific target promoters in trans, in particular those of mesodermal transcription factors [46]. A BAC-transgene expressing a single dose of Fendrr rescued the developmental defects caused by the loss of Fendrr, strongly suggesting that the observed phenotype is caused by lack of Fendrr RNA, and not by disruption of genomic sites or promoter elements of the adjacent Foxf1 gene. This conclusion was supported by a different knockout strategy, in which the first exon was left intact and which resulted in perinatal death [123]. In the early embryo, Fendrr is only transiently expressed in nascent lateral mesoderm [47]. However, later during development it is expressed in the lungs, a derivate of endoderm and lateral mesoderm, where it again might play an essential role [123, 128].
More lncRNAs involved in mesoderm and endoderm development have been reported. For instance DEANR1 (also known as ALIEN) is expressed downstream of the endoderm master regulator FOXA2 [76, 77]. DEANR1 depletion results in reduced expression of FOXA2 and accordingly to reduced differentiation of hESCs towards definitive endoderm. Interestingly, this defect can be rescued by restoring normal levels of FOXA2 expression, suggesting that DEANR1 functions mainly in cis. Another study described the same lncRNA, here called ALIEN, in cardiovascular progenitor cells in mouse and zebrafish. Depletion of ALIEN interfered with cardiac development, although the functional mechanism remains to be investigated further. DEANR1 was described to be localized in the nucleus, whereas ALIEN was seen in the perinuclear space and in the cytoplasm, even though both were analyzed in differentiating human stem cells. The discrepancy between the two studies might be a consequence of modifications in the differentiation method [76, 77].
Also, the transcription factor Pitx2, which is essential for the development of many different organs, especially during the establishment of left–right asymmetry, is regulated by a lncRNA in the embryonic gut [100]. In this case, the lncRNA Playrr is transcribed from a distant known enhancer region. The expression patterns of Pitx2 and Playrr are negatively correlated due to interference of Playrr with Pitx2 activation (for details see above).
LncRNAs expressed during neuronal and brain development have attracted particular attention. As described earlier, many lncRNAs are highly specifically expressed in distinct cell types, as illustratively shown in a targeted LacZ reporter screen [37] and in expression screens [36, 107, 129, 130]. A number of lncRNAs located close to the PouIII genes Pou3f3 and Pou3f2 (Brn1 and Brn2) encoding important neuronal transcription factors, were also found to play roles in neuronal development [37, 105, 123, 131]. The lncRNA Dali (or Dalir) (DNMT1-Associated Long Intergenic RNA) is transcribed 50 kb downstream of Pou3f3. The depletion of Dali leads to a reduction of Pou3f3 expression and to impaired neuronal differentiation [131]. Gene expression analysis upon knockdown and genome-wide localization (CHART) revealed POU3F3 dependent and independent targets of Dali. Furthermore, Dali physically interacts not only with various chromatin modifiers, but also with POU3F3 itself suggesting a multilayered association between Dali and POU3F3. The replacement of Pantr1 (POU domain, class 3, transcription factor 3 adjacent noncoding transcript), which is divergently transcribed to Pou3f3, by insertion of the lacZ gene did not influence Pou3f3 transcription itself, but modulated other Pou3f transcription factors in trans [37]. Pantr2 (or Linc-Brn1b), expressed convergent with Pou3f3, did not have this effect on Pou3f factors, and upon deletion led to fewer intermediate progenitors in the developing cortex. Thus, each of these three different lncRNAs, which are all transcribed near the Pou3f3 locus, affect neuronal development yet through distinct mechanisms.
Furthermore, another lncRNA located in a region important for neural development is Evf2 in the Dlx5/6 locus [69]. Evf2 was targeted by inserting a polyadenylation signal close to the promoter without deleting genomic DNA. Evf2 −/− embryos have less GABAergic neurons than wild type controls. This mutant effect could be rescued by in utero electroporation of Evf2 RNA, confirming that the phenotype is indeed dependent on this lncRNA.
RMST, also termed non-coding rhabdomyosarcoma (NCRMS) in humans, is a noncoding transcript expressed in rhabdomyosarcomas [132] and in developing dopaminergic neurons during murine development [133]. RMST is induced during neuronal differentiation of hESCs, and its depletion interferes with neuronal differentiation via its binding partner SOX2 (see above) [78, 107, 129]. Interestingly, RMST was found associated with obesity in a GWAS study, possibly pointing to a role of RMST in a neuronal regulatory network influencing energy balance [134].
Two more lncRNAs involved in nervous system development are the retinal noncoding RNA2 and 4 (RNCR2 and RNCR4). They were first found to be expressed in the developing retina and later also assigned a functional role in this process [135]. RNCR4 is part of a network including the RNA helicase DDX3X and the miR-183/96/182 that is essential for retinal architecture [136]. RNCR2, also known as MIAT or Gomafu, interferes with retinal cell differentiation when overexpressed or depleted [135, 137]. Furthermore MIAT is expressed earlier in the developing brain, where it localizes to the nuclear matrix [138]. Purifying distinct subpopulations of the developing brain at E14.5 revealed that MIAT is highly abundant in differentiating progenitors, where it is involved in corticogenesis by regulating the splicing pattern of Wnt7b [107] (Fig. 2g).
Another ectodermal derivative, the developing epidermis, has also been shown to be regulated by lncRNAs. During epidermal differentiation and maintenance, the nuclear lncRNA ANCR (anti-differentiation ncRNA) is essential for maintaining the proliferating progenitor pool by repressing differentiation factors, amongst them the transcription factors MAF and MAFB, probably in a PRC2 linked manner [139–141]. Depletion of ANCR leads to differentiation and loss of progenitor cells. Its differentiation promoting counterpart is a cytosolic lncRNA, terminal differentiation induced ncNRA (TINCR) that interacts with STAU1 and as a protein-RNA complex stabilizes mRNAs of pro-differentiation genes including MAF and MAFB [141, 142] (Fig. 2h). Ectopic expression of MAF:MAFB in TINCR depleted cells restored the expression of the differentiation gene signature showing that TINCR is crucial for ensuring appropriate protein levels in particular of MAF and MAFB.
Functions of lncRNAs in pathological settings
LncRNAs as a “new” class of molecules have also attracted attention in pathological settings. Here, we will highlight two very prominent areas, cancer and cardiovascular disease.
Cancer
Approximately one third of publications currently listed in PubMed linked with the keyword “lncRNA” also contain the keyword “cancer”. Even before the era of next generation sequencing, a few studies found lncRNAs to be dysregulated in tumors [35, 143, 144]. To date, numerous primary tumor samples, metastatic biopsies, the adjacent normal tissue and cancer cell lines have been profiled for their RNA expression and the results recapitulate the high degree of specificity found for noncoding RNAs in normal tissue compared to protein-coding genes [25, 67, 145–147]. Clustering of lncRNA expression patterns identifies subclasses of tumor types with high significance, opening the possibility to define tumor subtypes with a high chance to respond to targeted therapy. A study that considered samples from more than 5000 tumors from the cancer genome atlas (TCGA) project, demonstrated that, while approximately the same number of protein coding and lncRNA genes were dysregulated, 60 % of lncRNAs showed specificity for only one tumor type [146].
In addition, somatic copy number alterations (SCNA) found in cancer tissue often include lncRNA loci [57, 146, 147]. For example, the lncRNA FAL1, a lncRNA that was found amplified in a number of different tumor types, was shown to be important for the tumorigenic effects of different cell lines via a BMI1-mediated mechanism (see above), and its expression level and copy number significantly correlated with patient survival [57]. Furthermore, several lncRNAs have been shown to be prognostic for overall or disease-free survival of patients [67, 82, 148–150], information that is integrated into therapeutic decisions. Depletion of selected lncRNAs led to reduced malignancy, assayed as tumor size or metastatic potential in xenograft experiments [67, 72, 82, 101, 148, 149, 151–154], although these experiments were not able to distinguish between effects on tumor formation versus tumor progression. Further refinement was achieved when RNA knockdown was induced after tumor formation, mimicking therapeutic conditions. Reduced tumor growth was, for instance, observed upon FAL1 depletion [57]. In a different study, the inhibition of MALAT1 after tumor induction reduced metastasis formation in a xenograft lung cancer model and in a metastasizing breast cancer model [149, 155]. MALAT1 knockout mice do not show overt phenotypes [110, 156, 157] suggesting that down-regulation of MALAT1 in humans might also not be harmful for normal human cells, which makes it an attractive target for systemic application in patients.
A special case is the noncoding pseudo gene BRAFP1, which has been shown to be either mutated or aberrantly expressed in cancers, like its coding homologue BRAF. A mouse model revealed that induced overexpression of Braf-rs1 (the murine homologue of BRAFP1) leads to DLBCL (diffuse large B cell lymphomas)-like tumors. However, when the transgene was first induced to form tumors and then shut down again, the tumors regressed demonstrating the functional significance of the pseudo gene [158]. Braf-rs1 was shown to cause upregulation of Braf by acting as a ceRNA (competing endogenous RNA) and by “sponging” miRNAs that would otherwise target Braf transcripts.
One mechanism that cells can employ to prevent tumor formation is oncogene-induced senescence (OIS) [159]. The INK4B-ARF-INK4A locus, which plays a major role in OIS, is silenced in proliferating cells by Polycomb group proteins. This mechanism is dependent on the lncRNA ANRIL, which is transcribed from within the locus and acts in cis [58, 59, 160]. Furthermore, the MIR31HG gene located 400 kb upstream of the INK4B-ARF-INK4A locus, also encodes a lncRNA involved in the regulation of INK4A [161]. A third lncRNA functioning in OIS, UCA1, stabilizes mRNAs of protein-coding genes involved in the senescence pathway, probably by sequestering hnRNPKA1 [162]. Also, the tumor suppressor p53 and its associated signaling pathways include lncRNAs. p53 binds and regulates numerous lncRNAs, such as the linc-p21, which in turn regulates gene expression in concert with p53 and p21 [163–166].
Cardiovascular diseases
LncRNAs have also been linked to cardiovascular diseases (CVD), which are a major cause of death in western societies, and have been studied both in patient data, as well as in mouse models. It was observed that several genomic regions significantly associated with myocardial infarction encode lncRNAs, for example MIAT and ANRIL [167, 168]. Deletion of a 70 kb region including a large proportion of the ANRIL locus led to increased mortality in mice, particularly in combination with a high fat diet [169].
In other studies, massive parallel RNA sequencing revealed numerous lncRNAs displaying altered expression following surgically-induced myocardial infarction in mice [170, 171]. In cellular differentiation models, the depletion of selected lncRNAs impaired cardiomyocyte formation making them promising candidates for in vivo assessment of their functions [172]. Analysis of the noncoding transcripts originating from the myosin heavy chain 7 (Myh7) locus, an important structural protein required for heart contractions, revealed that one, the lncRNA Mhrt, was down-regulated after induced myocardial infarction. Ectopic expression of Mhrt in mice, which had been subjected to this operation reduced the severity of symptoms observed in control mice during recovery [68]. When lncRNAs in plasma from myocardial infarction patients were profiled, a mitochondrial lncRNA named long intergenic noncoding RNA predicting cardiac remodeling (LIPCAR) was found to significantly correlate with mortality caused by CVD and could thus be considered as a biomarker [173]. Whether this lncRNA also plays a causative role in infarction or heart failure remains to be investigated.
LncRNAs have been widely suggested as biomarkers and therapeutic targets in certain pathological settings. One reason for this is the extremely specific expression of many lncRNAs, opening the potential of targeting cellular subgroups rather than affecting patients systemically as in conventional treatments. Even if a lncRNA itself is not the disease driver, it can serve as drug target in cases where it acts as co-factor for a driver gene, which is more broadly expressed. Also, since lncRNAs often play quantitative roles rather than acting as switches in genetic programs, side effects of therapeutic treatments might be easier to control.
Concluding remarks
In recent years, the analysis of lncRNAs has highlighted the large diversity of cellular functions involving RNA molecules and has greatly expanded the catalog of functions previously attributed to noncoding RNAs. By now, we can conclude that noncoding RNAs play roles at all levels of gene control, including epigenetic mechanisms and nuclear organization, as well as in RNA processing, stability and translation. The recent discoveries of lncRNA functions provide good reasons for the notion that further investigation of lncRNAs bears high chances for the discovery of novel gene regulatory mechanisms in the future.
So far, only few lncRNAs playing an essential role in embryonic development, organogenesis or tissue homeostasis have been reported. Studies based on in vitro differentiation systems can provide important insights into the molecular modes of lncRNA function in local gene control or in regulatory networks. However, with regard to the roles of lncRNAs, for example in lineage decisions and cellular differentiation many findings are hampered by the lack of definitive proof in vivo. In particular, data derived from in vitro differentiated stem cells, whose genomic integrity and quality is hard to control, have to be interpreted cautiously. Also, the type of genetic change introduced into a lncRNA gene/locus for functional analysis (transcription stop, knockin, deletion etc.) needs to be considered carefully in order to draw the right conclusions [174–176]. Since lncRNAs do not only act as transcripts in cis or trans, but can also be side products of transcription affecting the expression of overlapping genes or accommodate regulatory DNA elements in their genomic loci, the analysis of their exact mode of action and functional roles in development and disease processes is more complex and difficult than that of protein-coding genes. It is now up to the responsibility of journal editors and reviewers to enforce the high standards of investigation this new and exciting field of research deserves.
References
Okazaki Y, Furuno M, Kasukawa T et al (2002) Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420:563–573. doi:10.1038/nature01266
Djebali S, Davis CA, Merkel A et al (2012) Landscape of transcription in human cells. Nature 489:101–108. doi:10.1038/nature11233
Derrien T, Johnson R, Bussotti G et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22:1775–1789. doi:10.1101/gr.132159.111
Cabili MN, Trapnell C, Goff L et al (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25:1915–1927. doi:10.1101/gad.17446611
Guo X, Gao L, Wang Y et al (2015) Advances in long noncoding RNAs: identification, structure prediction and function annotation. Brief Funct Genomics. doi:10.1093/bfgp/elv022
Harrow J, Frankish A, Gonzalez JM et al (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22:1760–1774. doi:10.1101/gr.135350.111
Ingolia NT, Lareau LF, Weissman JS (2011) Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 147:789–802. doi:10.1016/j.cell.2011.10.002
Ruiz-Orera J, Messeguer X, Subirana JA, Alba MM (2014) Long non-coding RNAs as a source of new peptides. elife 3:e03523. doi:10.7554/eLife.03523
Guttman M, Russell P, Ingolia NT et al (2013) Ribosome profiling provides evidence that large noncoding RNAs do not encode proteins. Cell 154:240–251. doi:10.1016/j.cell.2013.06.009
Bánfai B, Jia H, Khatun J et al (2012) Long noncoding RNAs are rarely translated in two human cell lines. Genome Res 22:1646–1657. doi:10.1101/gr.134767.111
Bazzini AA, Johnstone TG, Christiano R et al (2014) Identification of small ORFs in vertebrates using ribosome footprinting and evolutionary conservation. EMBO J. doi:10.1002/embj.201488411
Frith MC, Forrest AR, Nourbakhsh E et al (2006) The abundance of short proteins in the mammalian proteome. PLoS Genet 2:e52. doi:10.1371/journal.pgen.0020052
Anderson DM, Anderson KM, Chang C-L et al (2015) A micropeptide encoded by a putative long noncoding RNA regulates muscle performance. Cell 160:595–606. doi:10.1016/j.cell.2015.01.009
Pauli A, Norris ML, Valen E et al (2014) Toddler: an embryonic signal that promotes cell movement via Apelin receptors. Science 343:1248636. doi:10.1126/science.1248636
Yan Y, Cooper C, Hamedani MK et al (2015) The steroid receptor RNA activator protein (SRAP) controls cancer cell migration/motility. FEBS Lett 589:4010–4018. doi:10.1016/j.febslet.2015.11.007
Rupaimoole R, Lee J, Haemmerle M et al (2015) Long noncoding RNA ceruloplasmin promotes cancer growth by altering glycolysis. Cell Rep 13:2395–2402. doi:10.1016/j.celrep.2015.11.047
Novikova IV, Hennelly SP, Tung C-S, Sanbonmatsu KY (2013) Rise of the RNA machines: exploring the structure of long non-coding RNAs. J Mol Biol 425:3731–3746. doi:10.1016/j.jmb.2013.02.030
Weeks KM (2015) Review toward all RNA structures, concisely. Biopolymers 103:438–448. doi:10.1002/bip.22601
Smola MJ, Rice GM, Busan S et al (2015) Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis. Nat Protoc 10:1643–1669. doi:10.1038/nprot.2015.103
Spitale RC, Flynn RA, Zhang QC et al (2015) Structural imprints in vivo decode RNA regulatory mechanisms. Nature. doi:10.1038/nature14263
Guttman M, Amit I, Garber M et al (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227. doi:10.1038/nature07672
Sun M, Gadad SS, Kim D-S, Kraus WL (2015) Discovery, annotation, and functional analysis of long noncoding RNAs controlling Cell-cycle gene expression and proliferation in breast cancer cells. Mol Cell. doi:10.1016/j.molcel.2015.06.023
Marques AC, Hughes J, Graham B et al (2013) Chromatin signatures at transcriptional start sites separate two equally populated yet distinct classes of intergenic long noncoding RNAs. Genome Biol 14:R131. doi:10.1186/gb-2013-14-11-r131
ENCODE Project Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature 489:57–74. doi:10.1038/nature11247
Iyer MK, Niknafs YS, Malik R et al (2015) The landscape of long noncoding RNAs in the human transcriptome. Nat Genet 47:199–208. doi:10.1038/ng.3192
Hansen TB, Jensen TI, Clausen BH et al (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388. doi:10.1038/nature11993
Memczak S, Jens M, Elefsinioti A et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338. doi:10.1038/nature11928
Werner MS, Ruthenburg AJ (2015) Nuclear fractionation reveals thousands of chromatin-tethered noncoding RNAs adjacent to active genes. Cell Rep 12:1089–1098. doi:10.1016/j.celrep.2015.07.033
Herman RC, Williams JG, Penman S (1976) Message and non-message sequences adjacent to poly(A) in steady state heterogeneous nuclear RNA of HeLa cells. Cell 7:429–437
Yue F, Cheng Y, Breschi A et al (2014) A comparative encyclopedia of DNA elements in the mouse genome. Nature 515:355–364. doi:10.1038/nature13992
Haerty W, Ponting CP (2015) Unexpected selection to retain high GC content and splicing enhancers within exons of multiexonic lncRNA loci. RNA 21:333–346. doi:10.1261/rna.047324.114
Nitsche A, Rose D, Fasold M et al (2015) Comparison of splice sites reveals that long noncoding RNAs are evolutionarily well conserved. RNA 21:801–812. doi:10.1261/rna.046342.114
Ulitsky I, Shkumatava A, Jan CH et al (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147:1537–1550. doi:10.1016/j.cell.2011.11.055
Necsulea A, Soumillon M, Warnefors M et al (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505:635–640. doi:10.1038/nature12943
Diederichs S (2014) The four dimensions of noncoding RNA conservation. Trends Genet 30:121–123. doi:10.1016/j.tig.2014.01.004
Mercer TR, Dinger ME, Sunkin SM et al (2008) Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA 105:716–721. doi:10.1073/pnas.0706729105
Goff LA, Groff AF, Sauvageau M et al (2015) Spatiotemporal expression and transcriptional perturbations by long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA 112:6855–6862. doi:10.1073/pnas.1411263112
Grote P, Herrmann BG (2015) Long noncoding RNAs in organogenesis: making the difference. Trends Genet 31:329–335. doi:10.1016/j.tig.2015.02.002
Plath K, Fang J, Mlynarczyk-Evans SK et al (2003) Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131–135. doi:10.1126/science.1084274
da Rocha ST, Boeva V, Escamilla-Del-Arenal M et al (2014) Jarid2 Is implicated in the initial Xist-induced targeting of PRC2 to the inactive X chromosome. Mol Cell 53:301–316. doi:10.1016/j.molcel.2014.01.002
Zhao J, Sun BK, Erwin JA et al (2008) Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science 322:750–756. doi:10.1126/science.1163045
Rinn JL, Kertesz M, Wang JK et al (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129:1311–1323. doi:10.1016/j.cell.2007.05.022
Li L, Liu B, Wapinski OL et al (2013) Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep 5:3–12. doi:10.1016/j.celrep.2013.09.003
Schorderet P, Duboule D (2011) Structural and functional differences in the long non-coding RNA hotair in mouse and human. PLoS Genet 7:e1002071. doi:10.1371/journal.pgen.1002071
Klattenhoff CA, Scheuermann JC, Surface LE et al (2013) Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell 152:570–583. doi:10.1016/j.cell.2013.01.003
Grote P, Herrmann BG (2013) The long non-coding RNA Fendrr links epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol 10:1579–1585. doi:10.4161/rna.26165
Grote P, Wittler L, Hendrix D et al (2013) The tissue-specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24:206–214. doi:10.1016/j.devcel.2012.12.012
Marín-Béjar O, Marchese FP, Athie A et al (2013) Pint lincRNA connects the p53 pathway with epigenetic silencing by the Polycomb repressive complex 2. Genome Biol 14:R104. doi:10.1186/gb-2013-14-9-r104
Brockdorff N (2013) Noncoding RNA and Polycomb recruitment. RNA 19:429–442. doi:10.1261/rna.037598.112
Khalil AM, Guttman M, Huarte M et al (2009) Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106:11667–11672. doi:10.1073/pnas.0904715106
Guttman M, Donaghey J, Carey BW et al (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477:295–300. doi:10.1038/nature10398
Zhao J, Ohsumi TK, Kung JT et al (2010) Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell 40:939–953. doi:10.1016/j.molcel.2010.12.011
Schmitz K-M, Mayer C, Postepska A, Grummt I (2010) Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 24:2264–2269. doi:10.1101/gad.590910
O’Leary VB, Ovsepian SV, Carrascosa LG et al (2015) PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep 11:474–485. doi:10.1016/j.celrep.2015.03.043
Mondal T, Subhash S, Vaid R et al (2015) MEG3 long noncoding RNA regulates the TGF-β pathway genes through formation of RNA-DNA triplex structures. Nat Comms 6:7743. doi:10.1038/ncomms8743
Batista PJ, Chang HY (2013) Long noncoding RNAs: cellular address codes in development and disease. Cell 152:1298–1307. doi:10.1016/j.cell.2013.02.012
Hu X, Feng Y, Zhang D et al (2014) A functional genomic approach identifies FAL1 as an oncogenic long noncoding RNA that associates with BMI1 and represses p21 expression in cancer. Cancer Cell 26:344–357. doi:10.1016/j.ccr.2014.07.009
Pasmant E, Laurendeau I, Héron D et al (2007) Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res 67:3963–3969. doi:10.1158/0008-5472.CAN-06-2004
Yap KL, Li S, Muñoz-Cabello AM et al (2010) Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38:662–674. doi:10.1016/j.molcel.2010.03.021
Trievel RC, Shilatifard A (2009) WDR5, a complexed protein. Nat Struct Mol Biol 16:678–680. doi:10.1038/nsmb0709-678
Yang YW, Flynn RA, Chen Y et al (2014) Essential role of lncRNA binding for WDR5 maintenance of active chromatin and embryonic stem cell pluripotency. elife 3:e02046. doi:10.7554/eLife.02046
Wang KC, Yang YW, Liu B et al (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472:120–124. doi:10.1038/nature09819
Gomez JA, Wapinski OL, Yang YW et al (2013) The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-γ locus. Cell 152:743–754. doi:10.1016/j.cell.2013.01.015
Di Ruscio A, Ebralidze AK, Benoukraf T et al (2013) DNMT1-interacting RNAs block gene-specific DNA methylation. Nature 503:371–376. doi:10.1038/nature12598
Arab K, Park YJ, Lindroth AM et al (2014) Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell. doi:10.1016/j.molcel.2014.06.031
Hamazaki N, Uesaka M, Nakashima K et al (2015) Gene activation-associated long noncoding RNAs function in mouse preimplantation development. Development 142:910–920. doi:10.1242/dev.116996
Prensner JR, Iyer MK, Sahu A et al (2013) The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet 45:1392–1398. doi:10.1038/ng.2771
Han P, Li W, Lin C-H et al (2014) A long noncoding RNA protects the heart from pathological hypertrophy. Nature 514:102–106. doi:10.1038/nature13596
Bond AM, Vangompel MJW, Sametsky EA et al (2009) Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 12:1020–1027. doi:10.1038/nn.2371
Cajigas I, Leib DE, Cochrane J et al (2015) Evf2 lncRNA/BRG1/DLX1 interactions reveal RNA-dependent chromatin remodeling inhibition. Development 142:2641–2652. doi:10.1242/dev.126318
Minajigi A, Froberg JE, Wei C et al (2015) A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science 349:aab2276. doi:10.1126/science.aab2276
Wang Y, He L, Du Y et al (2015) The long noncoding RNA lncTCF7 promotes self-renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell 16:413–425. doi:10.1016/j.stem.2015.03.003
Kaneko S, Bonasio R, Saldaña-Meyer R et al (2013) Interactions between JARID2 and noncoding RNAs regulate PRC2 recruitment to chromatin. Mol Cell. doi:10.1016/j.molcel.2013.11.012
Merry CR, Forrest ME, Sabers JN et al (2015) DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum Mol Genet. doi:10.1093/hmg/ddv343
Davidovich C, Wang X, Cifuentes-Rojas C et al (2015) Toward a Consensus on the Binding Specificity and Promiscuity of PRC2 for RNA. Mol Cell. doi:10.1016/j.molcel.2014.12.017
Jiang W, Liu Y, Liu R et al (2015) The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep 11:137–148. doi:10.1016/j.celrep.2015.03.008
Kurian L, Aguirre A, Sancho-Martinez I et al (2015) Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 131:1278–1290. doi:10.1161/CIRCULATIONAHA.114.013303
Ng S-Y, Bogu GK, Soh BS, Stanton LW (2013) The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 51:349–359. doi:10.1016/j.molcel.2013.07.017
Rapicavoli NA, Qu K, Zhang J et al (2013) A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. elife 2:e00762. doi:10.7554/eLife.00762
Krawczyk M, Emerson BM (2014) p50-associated COX-2 extragenic RNA (PACER) activates COX-2 gene expression by occluding repressive NF-κB complexes. elife 3:e01776
Wang P, Xue Y, Han Y et al (2014) The STAT3-binding long noncoding RNA lnc-DC controls human dendritic cell differentiation. Science 344:310–313. doi:10.1126/science.1251456
Xing Z, Lin A, Li C et al (2014) lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell 159:1110–1125. doi:10.1016/j.cell.2014.10.013
Kung JT, Kesner B, An JY et al (2015) Locus-specific targeting to the X chromosome revealed by the RNA interactome of CTCF. Mol Cell. doi:10.1016/j.molcel.2014.12.006
Sun S, Del Rosario BC, Szanto A et al (2013) Jpx RNA activates Xist by evicting CTCF. Cell 153:1537–1551. doi:10.1016/j.cell.2013.05.028
Kim T-K, Hemberg M, Gray JM et al (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465:182–187. doi:10.1038/nature09033
Koch F, Fenouil R, Gut M et al (2011) Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol 18:956–963. doi:10.1038/nsmb.2085
Natoli G, Andrau J-C (2012) Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet 46:1–19. doi:10.1146/annurev-genet-110711-155459
Collis P, Antoniou M, Grosveld F (1990) Definition of the minimal requirements within the human beta-globin gene and the dominant control region for high level expression. EMBO J 9:233–240
Ashe HL, Monks J, Wijgerde M et al (1997) Intergenic transcription and transinduction of the human beta-globin locus. Genes Dev 11:2494–2509
Orom UA, Shiekhattar R (2011) Long non-coding RNAs and enhancers. Curr Opin Genet Dev 21:194–198. doi:10.1016/j.gde.2011.01.020
Andersson R, Gebhard C, Miguel-Escalada I et al (2014) An atlas of active enhancers across human cell types and tissues. Nature 507:455–461. doi:10.1038/nature12787
Ounzain S, Pezzuto I, Micheletti R et al (2014) Functional importance of cardiac enhancer-associated noncoding RNAs in heart development and disease. J Mol Cell Cardiol 76:55–70. doi:10.1016/j.yjmcc.2014.08.009
Orom UA, Derrien T, Beringer M et al (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46–58. doi:10.1016/j.cell.2010.09.001
Li W, Notani D, Ma Q et al (2013) Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498:516–520. doi:10.1038/nature12210
Lai F, Orom UA, Cesaroni M et al (2013) Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494:497–501. doi:10.1038/nature11884
Schaukowitch K, Joo J-Y, Liu X et al (2014) Enhancer RNA facilitates NELF release from immediate early genes. Mol Cell 56:29–42. doi:10.1016/j.molcel.2014.08.023
Mousavi K, Zare H, Dell’orso S et al (2013) eRNAs promote transcription by establishing chromatin accessibility at defined genomic loci. Mol Cell 51:606–617. doi:10.1016/j.molcel.2013.07.022
Yin Y, Yan P, Lu J et al (2015) Opposing roles for the lncRNA haunt and its genomic locus in regulating HOXA Gene activation during embryonic stem cell differentiation. Cell Stem Cell 16:504–516. doi:10.1016/j.stem.2015.03.007
Maamar H, Cabili MN, Rinn J, Raj A (2013) linc-HOXA1 is a noncoding RNA that represses Hoxa1 transcription in cis. Genes Dev 27:1260–1271. doi:10.1101/gad.217018.113
Welsh IC, Kwak H, Chen FL et al (2015) Chromatin architecture of the Pitx2 locus requires CTCF- and Pitx2-dependent asymmetry that mirrors embryonic gut laterality. Cell Rep. doi:10.1016/j.celrep.2015.08.075
Yang L, Lin C, Jin C et al (2013) lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature 500:598–602. doi:10.1038/nature12451
Yang F, Deng X, Ma W et al (2015) The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol 16:52. doi:10.1186/s13059-015-0618-0
Hacisuleyman E, Goff LA, Trapnell C et al (2014) Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21:198–206. doi:10.1038/nsmb.2764
Bergmann JH, Li J, Eckersley-Maslin MA et al (2015) Regulation of the ESC transcriptome by nuclear long non-coding RNAs. Genome Res 25:1336–1346. doi:10.1101/gr.189027.114
Ramos AD, Andersen RE, Liu SJ et al (2015) The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16:439–447. doi:10.1016/j.stem.2015.02.007
Lin N, Chang K-Y, Li Z et al (2014) An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 53:1005–1019. doi:10.1016/j.molcel.2014.01.021
Aprea J, Prenninger S, Dori M et al (2013) Transcriptome sequencing during mouse brain development identifies long non-coding RNAs functionally involved in neurogenic commitment. EMBO J 32:3145–3160. doi:10.1038/emboj.2013.245
Barry G, Briggs JA, Vanichkina DP et al (2014) The long non-coding RNA Gomafu is acutely regulated in response to neuronal activation and involved in schizophrenia-associated alternative splicing. Mol Psychiatry 19:486–494. doi:10.1038/mp.2013.45
Tripathi V, Ellis JD, Shen Z et al (2010) The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39:925–938. doi:10.1016/j.molcel.2010.08.011
Eißmann M, Gutschner T, Hämmerle M et al (2012) Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol 9:1076–1087. doi:10.4161/rna.21089
Lee S, Kopp F, Chang T-C et al (2015) Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. doi:10.1016/j.cell.2015.12.017
Chen CY, Sarnow P (1995) Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268:415–417. doi:10.1126/science.7536344
Yan L, Yang M, Guo H et al (2013) Single-cell RNA-Seq profiling of human preimplantation embryos and embryonic stem cells. Nat Struct Mol Biol 20:1131–1139. doi:10.1038/nsmb.2660
Brannan CI, Dees EC, Ingram RS, Tilghman SM (1990) The product of the H19 gene may function as an RNA. Mol Cell Biol 10:28–36
Bartolomei MS, Zemel S, Tilghman SM (1991) Parental imprinting of the mouse H19 gene. Nature 351:153–155. doi:10.1038/351153a0
Leighton PA, Ingram RS, Eggenschwiler J et al (1995) Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375:34–39. doi:10.1038/375034a0
Dey BK, Pfeifer K, Dutta A (2014) The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration. Genes Dev 28:491–501. doi:10.1101/gad.234419.113
Latos PA, Pauler FM, Koerner MV et al (2012) Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338:1469–1472. doi:10.1126/science.1228110
Kanduri C (2016) Long noncoding RNAs: lessons from genomic imprinting. Biochim Biophys Acta 1859:102–111. doi:10.1016/j.bbagrm.2015.05.006
Penny GD, Kay GF, Sheardown SA et al (1996) Requirement for Xist in X chromosome inactivation. Nature 379:131–137. doi:10.1038/379131a0
Marahrens Y, Panning B, Dausman J et al (1997) Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11:156–166
Zhou Y, Cheunsuchon P, Nakayama Y et al (2010) Activation of paternally expressed genes and perinatal death caused by deletion of the Gtl2 gene. Development 137:2643–2652. doi:10.1242/dev.045724
Sauvageau M, Goff LA, Lodato S et al (2013) Multiple knockout mouse models reveal lincRNAs are required for life and brain development. elife 2:e01749. doi:10.7554/eLife.01749
Nakagawa S, Naganuma T, Shioi G, Hirose T (2011) Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol 193:31–39. doi:10.1083/jcb.201011110
Oliver PL, Chodroff RA, Gosal A et al (2015) Disruption of Visc-2, a brain-expressed conserved long noncoding RNA, does not elicit an overt anatomical or behavioral phenotype. Cereb Cortex 25:3572–3585. doi:10.1093/cercor/bhu196
Li L, Chang HY (2014) Physiological roles of long noncoding RNAs: insight from knockout mice. Trends Cell Biol. doi:10.1016/j.tcb.2014.06.003
Nakagawa S, Shimada M, Yanaka K et al (2014) The lncRNA Neat1 is required for corpus luteum formation and the establishment of pregnancy in a subpopulation of mice. Development. doi:10.1242/dev.110544
Herriges MJ, Swarr DT, Morley MP et al (2014) Long noncoding RNAs are spatially correlated with transcription factors and regulate lung development. Genes Dev 28:1363–1379. doi:10.1101/gad.238782.114
Ng S-Y, Johnson R, Stanton LW (2012) Human long non-coding RNAs promote pluripotency and neuronal differentiation by association with chromatin modifiers and transcription factors. EMBO J 31:522–533. doi:10.1038/emboj.2011.459
Ramos AD, Diaz A, Nellore A et al (2013) Integration of genome-wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 12:616–628. doi:10.1016/j.stem.2013.03.003
Chalei V, Sansom SN, Kong L et al (2014) The long non-coding RNA Dali is an epigenetic regulator of neural differentiation. elife 3:e04530. doi:10.7554/eLife.04530
Chan AS, Thorner PS, Squire JA, Zielenska M (2002) Identification of a novel gene NCRMS on chromosome 12q21 with differential expression between rhabdomyosarcoma subtypes. Oncogene 21:3029–3037. doi:10.1038/sj.onc.1205460
Uhde CW, Vives J, Jaeger I, Li M (2010) Rmst is a novel marker for the mouse ventral mesencephalic floor plate and the anterior dorsal midline cells. PLoS One 5:e8641. doi:10.1371/journal.pone.0008641
Wheeler E, Huang N, Bochukova EG et al (2013) Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat Genet 45:513–517. doi:10.1038/ng.2607
Blackshaw S, Harpavat S, Trimarchi J et al (2004) Genomic analysis of mouse retinal development. PLoS Biol 2:E247. doi:10.1371/journal.pbio.0020247
Krol J, Krol I, Alvarez CPP et al (2015) A network comprising short and long noncoding RNAs and RNA helicase controls mouse retina architecture. Nat Comms 6:7305. doi:10.1038/ncomms8305
Rapicavoli NA, Poth EM, Blackshaw S (2010) The long noncoding RNA RNCR2 directs mouse retinal cell specification. BMC Dev Biol 10:49. doi:10.1186/1471-213X-10-49
Sone M, Hayashi T, Tarui H et al (2007) The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci 120:2498–2506. doi:10.1242/jcs.009357
Kretz M, Webster DE, Flockhart RJ et al (2012) Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 26:338–343. doi:10.1101/gad.182121.111
Zhu L, Xu P-C (2013) Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem Biophys Res Commun 432:612–617. doi:10.1016/j.bbrc.2013.02.036
Lopez-Pajares V, Qu K, Zhang J et al (2015) A LncRNA-MAF:MAFB transcription factor network regulates epidermal differentiation. Dev Cell 32:693–706. doi:10.1016/j.devcel.2015.01.028
Kretz M, Siprashvili Z, Chu C et al (2013) Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 493:231–235. doi:10.1038/nature11661
Ji P, Diederichs S, Wang W et al (2003) MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22:8031–8041. doi:10.1038/sj.onc.1206928
Srikantan V, Zou Z, Petrovics G et al (2000) PCGEM1, a prostate-specific gene, is overexpressed in prostate cancer. Proc Natl Acad Sci USA 97:12216–12221. doi:10.1073/pnas.97.22.12216
Brunner AL, Beck AH, Edris B et al (2012) Transcriptional profiling of long non-coding RNAs and novel transcribed regions across a diverse panel of archived human cancers. Genome Biol 13:R75. doi:10.1186/gb-2012-13-8-r75
Yan X, Hu Z, Feng Y et al (2015) Comprehensive genomic characterization of long non-coding RNAs across human cancers. Cancer Cell 28:529–540. doi:10.1016/j.ccell.2015.09.006
Du Z, Fei T, Verhaak RGW et al (2013) Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol 20:908–913. doi:10.1038/nsmb.2591
Pandey GK, Mitra S, Subhash S et al (2014) The risk-associated long noncoding RNA NBAT-1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 26:722–737. doi:10.1016/j.ccell.2014.09.014
Gutschner T, Hämmerle M, Eißmann M et al (2013) The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 73:1180–1189. doi:10.1158/0008-5472.CAN-12-2850
Xue S, Li Q-W, Che J-P et al (2015) Decreased expression of long non-coding RNA NBAT-1 is associated with poor prognosis in patients with clear cell renal cell carcinoma. Int J Clin Exp Pathol 8:3765–3774
Trimarchi T, Bilal E, Ntziachristos P et al (2014) Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell 158:593–606. doi:10.1016/j.cell.2014.05.049
Gupta RA, Shah N, Wang KC et al (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076. doi:10.1038/nature08975
Liu B, Sun L, Liu Q et al (2015) A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 27:370–381. doi:10.1016/j.ccell.2015.02.004
Dijkstra JM, Alexander DB (2015) The “NF-κB interacting long noncoding RNA” (NKILA) transcript is antisense to cancer-associated gene PMEPA1. F1000Res 4:96. doi:10.12688/f1000research.6400.1
Arun G, Diermeier S, Akerman M et al (2015) Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 30:34–51. doi:10.1101/gad.270959.115
Nakagawa S, Ip JY, Shioi G et al (2012) Malat1 is not an essential component of nuclear speckles in mice. RNA 18:1487–1499. doi:10.1261/rna.033217.112
Zhang B, Arun G, Mao YS et al (2012) The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2:111–123. doi:10.1016/j.celrep.2012.06.003
Karreth FA, Reschke M, Ruocco A et al (2015) The BRAF pseudogene functions as a competitive endogenous RNA and induces lymphoma in vivo. Cell. doi:10.1016/j.cell.2015.02.043
Gil J, Peters G (2006) Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat Rev Mol Cell Biol 7:667–677. doi:10.1038/nrm1987
Wan G, Mathur R, Hu X et al (2013) Long non-coding RNA ANRIL (CDKN2B-AS) is induced by the ATM-E2F1 signaling pathway. Cell Signal 25:1086–1095. doi:10.1016/j.cellsig.2013.02.006
Montes M, Nielsen MM, Maglieri G et al (2015) The lncRNA MIR31HG regulates p16(INK4A) expression to modulate senescence. Nat Comms 6:6967. doi:10.1038/ncomms7967
Kumar PP, Emechebe U, Smith R et al (2014) Coordinated control of senescence by lncRNA and a novel T-box3 co-repressor complex. elife. doi:10.7554/eLife.02805
Huarte M, Guttman M, Feldser D et al (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142:409–419. doi:10.1016/j.cell.2010.06.040
Sánchez Y, Segura V, Marín-Béjar O et al (2014) Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Comms 5:5812. doi:10.1038/ncomms6812
Dimitrova N, Zamudio JR, Jong RM et al (2014) LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 54:777–790. doi:10.1016/j.molcel.2014.04.025
Wu G, Cai J, Han Y et al (2014) LincRNA-p21 regulates neointima formation, vascular smooth muscle cell proliferation, apoptosis, and atherosclerosis by enhancing p53 activity. Circulation 130:1452–1465. doi:10.1161/CIRCULATIONAHA.114.011675
Ishii N, Ozaki K, Sato H et al (2006) Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J Hum Genet 51:1087–1099. doi:10.1007/s10038-006-0070-9
Broadbent HM, Peden JF, Lorkowski S et al (2008) Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet 17:806–814. doi:10.1093/hmg/ddm352
Visel A, Zhu Y, May D et al (2010) Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature 464:409–412. doi:10.1038/nature08801
Matkovich SJ, Edwards JR, Grossenheider TC et al (2014) Epigenetic coordination of embryonic heart transcription by dynamically regulated long noncoding RNAs. Proc Natl Acad Sci USA 111:12264–12269. doi:10.1073/pnas.1410622111
Ounzain S, Micheletti R, Beckmann T et al (2015) Genome-wide profiling of the cardiac transcriptome after myocardial infarction identifies novel heart-specific long non-coding RNAs. Eur Heart J 36:353–368a. doi:10.1093/eurheartj/ehu180
Ounzain S, Micheletti R, Arnan C et al (2015) CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol. doi:10.1016/j.yjmcc.2015.09.016
Kumarswamy R, Bauters C, Volkmann I et al (2014) Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res 114:1569–1575. doi:10.1161/CIRCRESAHA.114.303915
Bassett AR, Akhtar A, Barlow DP et al (2014) Considerations when investigating lncRNA function in vivo. elife 3:e03058
Janowski BA, Huffman KE, Schwartz JC et al (2005) Inhibiting gene expression at transcription start sites in chromosomal DNA with antigene RNAs. Nat Chem Biol 1:216–222. doi:10.1038/nchembio725
Goff LA, Rinn JL (2015) Linking RNA biology to lncRNAs. Genome Res 25:1456–1465. doi:10.1101/gr.191122.115
Stadtfeld M, Apostolou E, Ferrari F, et al. (2012) Ascorbic acid prevents loss of Dlk1-Dio3 imprinting and facilitates generation of all-iPS cell mice from terminally differentiated B cells. Nat Genet 44:398–405, S1–S2. doi:10.1038/ng.1110
Zhang X, Lian Z, Padden C et al (2009) A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 113:2526–2534. doi:10.1182/blood-2008-06-162164
Delpretti S, Montavon T, Leleu M et al (2013) Multiple enhancers regulate Hoxd genes and the Hotdog LncRNA during cecum budding. Cell Rep. doi:10.1016/j.celrep.2013.09.002
Gong C, Li Z, Ramanujan K et al (2015) A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Dev Cell 34:181–191. doi:10.1016/j.devcel.2015.05.009
Prensner JR, Sahu A, Iyer MK et al (2014) The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget 5:1434–1438
McHugh CA, Chen C-K, Chow A et al (2015) The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature 521:232–236. doi:10.1038/nature14443
Kay GF, Penny GD, Patel D et al (1993) Expression of Xist during mouse development suggests a role in the initiation of X chromosome inactivation. Cell 72:171–182
Acknowledgments
We thank Arica M Beisaw, Jesse Veenvliet, and Pavel Tsaytler for valuable comments on the manuscript. The authors’ work is funded by the Max Planck Society (B.G.H.; S.U.S) and the DFG (German Research Foundation) Excellence Cluster Cardio-Pulmonary System (Exc147-2) (P.G.).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Schmitz, S.U., Grote, P. & Herrmann, B.G. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 73, 2491–2509 (2016). https://doi.org/10.1007/s00018-016-2174-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-016-2174-5