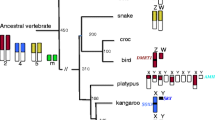
Abstract
Ohno’s hypothesis states that dosage compensation in mammals evolved in two steps: a twofold hyperactivation of the X chromosome in both sexes to compensate for gene losses on the Y chromosome, and silencing of one X (X-chromosome inactivation, XCI) in females to restore optimal dosage. Recent tests of this hypothesis have returned contradictory results. In this review, we explain this ongoing controversy and argue that a novel view on dosage compensation evolution in mammals is starting to emerge. Ohno’s hypothesis may be true for a few, dosage-sensitive genes only. If so few genes are compensated, then why has XCI evolved as a chromosome-wide mechanism? This and several other questions raised by the new data in mammals are discussed, and future research directions are proposed.



Similar content being viewed by others
Abbreviations
- XCI:
-
X-chromosome inactivation
- PAR:
-
Pseudoautosomal region
- rXCI:
-
Random X-chromosome inactivation
- pXCI:
-
Paternal X-chromosome inactivation
- Xi:
-
Inactivated X chromosome
- PolII:
-
RNA polymerase II
- ZGA:
-
Zygote genome activation
- NGS:
-
Next-generation sequencing
- Ne:
-
Effective population size
- PAM:
-
Parental antagonism model
- XIC:
-
X-inactivation center
References
Veyrunes F, Waters PD, Miethke P, Rens W, McMillan D, Alsop AE, Grutzner F, Deakin JE, Whittington CM, Schatzkamer K, Kremitzki CL, Graves T, Ferguson-Smith MA, Warren W, Marshall Graves JA (2008) Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res 18(6):965–973. doi:10.1101/gr.7101908
Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H (2008) Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol 6(4):e80. doi:10.1371/journal.pbio.0060080
Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou S-F, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang S-P, Waterston RH, Wilson RK, Rozen S, Page DC (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423:825–837
Ohno S, Kaplan WD, Kinosita R (1959) Formation of the sex chromatin by a single X-chromosome in liver cells of Rattus norvegicus. Exp Cell Res 18(2):415–419
Lyon MF (1961) Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190:372–373
Ohno S (1967) Sex chromosomes and sex linked genes. Springer, Berlin Heidelberg New York
Deakin JE, Chaumeil J, Hore TA, Marshall Graves JA (2009) Unravelling the evolutionary origins of X chromosome inactivation in mammals: insights from marsupials and monotremes. Chromosome Res 17(5):671–685. doi:10.1007/s10577-009-9058-6
Lee JT (2011) Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat Rev Mol Cell Biol 12(12):815–826. doi:10.1038/nrm3231
Livernois AM, Graves JA, Waters PD (2012) The origin and evolution of vertebrate sex chromosomes and dosage compensation. Heredity (Edinb) 108(1):50–58. doi:10.1038/hdy.2011.106
Jeon Y, Sarma K, Lee JT (2012) New and Xisting regulatory mechanisms of X chromosome inactivation. Curr Opin Genet Dev 22(2):62–71. doi:10.1016/j.gde.2012.02.007
Disteche CM (2012) Dosage compensation of the sex chromosomes. Annu Rev Genet 46:537–560. doi:10.1146/annurev-genet-110711-155454
Gribnau J, Grootegoed JA (2012) Origin and evolution of X chromosome inactivation. Curr Opin Cell Biol 24(3):397–404. doi:10.1016/j.ceb.2012.02.004
Dupont C, Gribnau J (2013) Different flavors of X-chromosome inactivation in mammals. Curr Opin Cell Biol 25(3):314–321. doi:10.1016/j.ceb.2013.03.001
Ohhata T, Wutz A (2013) Reactivation of the inactive X chromosome in development and reprogramming. Cell Mol Life Sci 70(14):2443–2461. doi:10.1007/s00018-012-1174-3
Schulz EG, Heard E (2013) Role and control of X chromosome dosage in mammalian development. Curr Opin Genet Dev 23(2):109–115. doi:10.1016/j.gde.2013.01.008
Gontan C, Achame EM, Demmers J, Barakat TS, Rentmeester E, van Ijcken W, Grootegoed JA, Gribnau J (2012) RNF12 initiates X-chromosome inactivation by targeting REX1 for degradation. Nature 485(7398):386–390. doi:10.1038/nature11070
Engreitz JM, Pandya-Jones A, McDonel P, Shishkin A, Sirokman K, Surka C, Kadri S, Xing J, Goren A, Lander ES, Plath K, Guttman M (2013) The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341(6147):1237973. doi:10.1126/science.1237973
Chow JC, Ciaudo C, Fazzari MJ, Mise N, Servant N, Glass JL, Attreed M, Avner P, Wutz A, Barillot E, Greally JM, Voinnet O, Heard E (2010) LINE-1 activity in facultative heterochromatin formation during X chromosome inactivation. Cell 141(6):956–969. doi:10.1016/j.cell.2010.04.042
Carrel L, Willard HF (2005) X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature 434:400–404
Zhang Y, Morales AC, Jiang M, Zhu Y, Hu L, Urrutia AO, Kong X, Hurst LD (2013) Genes that escape X-inactivation in humans have high intraspecific variability in expression, are associated with mental impairment but are not slow evolving. Mol Biol Evol. doi:10.1093/molbev/mst148
Duret L, Chureau C, Samain S, Weissenbach J, Avner P (2006) The Xist RNA gene evolved in eutherians by pseudogenization of a protein-coding gene. Science 312:1653–1655
Nguyen DK, Disteche CM (2006) Dosage compensation of the active X chromosome in mammals. Nat Genet 38(1):47–53
Gupta V, Parisi M, Sturgill D, Nuttall R, Doctolero M, Dudko OK, Malley JD, Eastman PS, Oliver B (2006) Global analysis of X-chromosome dosage compensation. J. Biol 5(1):3. doi:10.1186/jbiol30
Talebizadeh Z, Simon SD, Butler MG (2006) X chromosome gene expression in human tissues: male and female comparisons. Genomics 88(6):675–681. doi:10.1016/j.ygeno.2006.07.016
Lin H, Gupta V, Vermilyea MD, Falciani F, Lee JT, O’Neill LP, Turner BM (2007) Dosage compensation in the mouse balances up-regulation and silencing of X-linked genes. PLoS Biol 5(12):e326. doi:10.1371/journal.pbio.0050326
Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT (2008) Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet 4(1):e9. doi:10.1371/journal.pgen.0040009
Xiong Y, Chen X, Chen Z, Wang X, Shi S, Zhang J, He X (2010) RNA sequencing shows no dosage compensation of the active X-chromosome. Nat Genet 42(12):1043–1047
Deng X, Hiatt JB, Nguyen DK, Ercan S, Sturgill D, Hillier LW, Schlesinger F, Davis CA, Reinke VJ, Gingeras TR, Shendure J, Waterston RH, Oliver B, Lieb JD, Disteche CM (2011) Evidence for compensatory upregulation of expressed X-linked genes in mammals, Caenorhabditis elegans and Drosophila melanogaster. Nat Genet 43(12):1179–1185. doi:10.1038/ng.948
He X, Chen X, Xiong Y, Chen Z, Wang X, Shi S, Wang X, Zhang J (2011) He et al. reply. Nat Genet 43(12):1171–1172
Kharchenko PV, Xi R, Park PJ (2011) Evidence for dosage compensation between the X chromosome and autosomes in mammals. Nat Genet 43(12):1167–1169. doi:10.1038/ng.991 (author reply 1171-1162)
Lin H, Halsall JA, Antczak P, O’Neill LP, Falciani F, Turner BM (2011) Relative overexpression of X-linked genes in mouse embryonic stem cells is consistent with Ohno’s hypothesis. Nat Genet 43(12):1169–1170. doi:10.1038/ng.992 (author reply 1171-1162)
Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA (2012) Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci USA 109(14):5346–5351. doi:10.1073/pnas.1116763109
Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schutz F, Daish T, Grutzner F, Kaessmann H (2012) Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol 10(5):e1001328. doi:10.1371/journal.pbio.1001328
Lin F, Xing K, Zhang J, He X (2012) Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc Natl Acad Sci USA 109(29):11752–11757. doi:10.1073/pnas.1201816109
Yildirim E, Sadreyev RI, Pinter SF, Lee JT (2011) X-chromosome hyperactivation in mammals via nonlinear relationships between chromatin states and transcription. Nat Struct Mol Biol 19(1):56–61. doi:10.1038/nsmb.2195
Adler DA, Rugarli EI, Lingenfelter PA, Tsuchiya K, Poslinski D, Liggitt HD, Chapman VM, Elliott RW, Ballabio A, Disteche CM (1997) Evidence of evolutionary up-regulation of the single active X chromosome in mammals based on Clc4 expression levels in Mus spretus and Mus musculus. Proc Natl Acad Sci USA 94(17):9244–9248
Brawand D, Soumillon M, Necsulea A, Julien P, Csardi G, Harrigan P, Weier M, Liechti A, Aximu-Petri A, Kircher M, Albert FW, Zeller U, Khaitovich P, Grutzner F, Bergmann S, Nielsen R, Paabo S, Kaessmann H (2011) The evolution of gene expression levels in mammalian organs. Nature 478(7369):343–348. doi:10.1038/nature10532
Zhang YE, Vibranovski MD, Landback P, Marais GA, Long M (2010) Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol 8(10). doi:10.1371/journal.pbio.1000494
Malone JH, Cho DY, Mattiuzzo NR, Artieri CG, Jiang L, Dale RK, Smith HE, McDaniel J, Munro S, Salit M, Andrews J, Przytycka TM, Oliver B (2012) Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol 13(4):r28. doi:10.1186/gb-2012-13-4-r28
Birchler JA (2012) Claims and counterclaims of X-chromosome compensation. Nat Struct Mol Biol 19(1):3–5. doi:10.1038/nsmb.2218
Papp B, Pal C, Hurst LD (2003) Dosage sensitivity and the evolution of gene families in yeast. Nature 424(6945):194–197. doi:10.1038/nature01771
Hall DW, Wayne ML (2013) Ohno’s “peril of hemizygosity” revisited: gene loss, dosage compensation, and mutation. Genome Biol Evol 5(1):1–15. doi:10.1093/gbe/evs106
Mank JE, Hosken DJ, Wedell N (2011) Some inconvenient truths about sex chromosome dosage compensation and the potential role of sexual conflict. Evolution 65(8):2133–2144. doi:10.1111/j.1558-5646.2011.01316.x
Wright AE, Mank JE (2012) Battle of the sexes: conflict over dosage-sensitive genes and the origin of X chromosome inactivation. Proc Natl Acad Sci USA 109(14):5144–5145. doi:10.1073/pnas.1202905109
Veitia RA (2005) Gene dosage balance: deletions, duplications and dominance. Trends Genet 21(1):33–35. doi:10.1016/j.tig.2004.11.002
Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G (2005) Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics 169:1915–1925
Gout J, Kahn D, Duret L (2010) The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet 6(5):e1000944. doi:10.1371/journal.pgen.1000944
Engelstadter J, Haig D (2008) Sexual antagonism and the evolution of X chromosome inactivation. Evolution 62(8):2097–2104. doi:10.1111/j.1558-5646.2008.00431.x
Haig D (2000) The kinship theory of genomic imprinting. Annu Rev Ecol Syst 31:9–32
Haig D (2006) Self-imposed silence: parental antagonism and the evolution of X-chromosome inactivation. Evolution 60(3):440–447
Haig D (2006) Intragenomic politics. Cytogenet Genome Res 113(1–4):68–74. doi:10.1159/000090816
Necsulea A, Soumillon M, Liechti A, Daish T, Baker JC, Grützner F, Kaessmann H (2013) Functionality and evolution of lncRNA repertoires and expression patterns in tetrapods. Nature (in press)
Livernois AM, Waters SA, Deakin JE, Marshall Graves JA, Waters PD (2013) Independent evolution of transcriptional inactivation on sex chromosomes in birds and mammals. PLoS Genet 9(7):e1003635. doi:10.1371/journal.pgen.1003635
Collignon J, Sockanathan S, Hacker A, Cohen-Tannoudji M, Norris D, Rastan S, Stevanovic M, Goodfellow PN, Lovell-Badge R (1996) A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development 122(2):509–520
Splinter E, de Wit E, Nora EP, Klous P, van de Werken HJ, Zhu Y, Kaaij LJ, van Ijcken W, Gribnau J, Heard E, de Laat W (2011) The inactive X chromosome adopts a unique three-dimensional conformation that is dependent on Xist RNA. Genes Dev 25(13):1371–1383. doi:10.1101/gad.633311
Charlesworth B, Charlesworth D (1978) A model for the evolution of dioecy and gynodioecy. Amer Nat 112:975–997
Bachtrog D (2013) Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14(2):113–124. doi:10.1038/nrg3366
Rice WR (1987) The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex-chromosomes. Evolution 41:911–914
Charlesworth D, Charlesworth B, Marais G (2005) Steps in the evolution of heteromorphic sex chromosomes. Heredity 95(2):118–128
Grant J, Mahadevaiah SK, Khil P, Sangrithi MN, Royo H, Duckworth J, McCarrey JR, VandeBerg JL, Renfree MB, Taylor W, Elgar G, Camerini-Otero RD, Gilchrist MJ, Turner JM (2012) Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487(7406):254–258. doi:10.1038/nature11171
Wang X, Douglas KC, Vandeberg JL, Clark A, Samollow PB (2013) Chromosome-wide profiling of X-chromosome inactivation and epigenetic states in fetal brain and placenta of the opossum, Monodelphis domestica. Genome Res. doi:10.1101/gr.161919.113
Okamoto I, Patrat C, Thepot D, Peynot N, Fauque P, Daniel N, Diabangouaya P, Wolf JP, Renard JP, Duranthon V, Heard E (2011) Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 472(7343):370–374. doi:10.1038/nature09872
Tachibana M, Ma H, Sparman ML, Lee HS, Ramsey CM, Woodward JS, Sritanaudomchai H, Masterson KR, Wolff EE, Jia Y, Mitalipov SM (2012) X-chromosome inactivation in monkey embryos and pluripotent stem cells. Dev Biol 371(2):146–155. doi:10.1016/j.ydbio.2012.08.009
Wang X, Miller DC, Clark AG, Antczak DF (2012) Random X inactivation in the mule and horse placenta. Genome Res 22(10):1855–1863. doi:10.1101/gr.138487.112
Wang X, Soloway PD, Clark AG (2010) Paternally biased X inactivation in mouse neonatal brain. Genome Biol 11(7):R79. doi:10.1186/gb-2010-11-7-r79
Connallon T, Clark AG (2013) Sex-differential selection and the evolution of X inactivation strategies. PLoS Genet 9(4):e1003440. doi:10.1371/journal.pgen.1003440
Veyrunes F, Chevret P, Catalan J, Castiglia R, Watson J, Dobigny G, Robinson TJ, Britton-Davidian J (2010) A novel sex determination system in a close relative of the house mouse. Proc Biol Sci 277(1684):1049–1056. doi:10.1098/rspb 2009.1925
Castagne R, Rotival M, Zeller T, Wild PS, Truong V, Tregouet DA, Munzel T, Ziegler A, Cambien F, Blankenberg S, Tiret L (2011) The choice of the filtering method in microarrays affects the inference regarding dosage compensation of the active X-chromosome. PLoS ONE 6(9):e23956. doi:10.1371/journal.pone.0023956
Jue NK, Murphy MB, Kasowitz SD, Qureshi SM, Obergfell CJ, Elsisi S, Foley RJ, O’Neill RJ, O’Neill MJ (2013) Determination of dosage compensation of the mammalian X chromosome by RNA-seq is dependent on analytical approach. BMC Genomics 14(1):150. doi:10.1186/1471-2164-14-150
Mank JE, Ellegren H (2009) Sex bias in gene expression is not the same as dosage compensation. Heredity 103(5):434
Trivers RL (1972) Parental investment and sexual selection. In: Campbell B (ed) Sexual selection and the descent of man 1871-1971. Aldine-Atherton, Chicago, pp 136–179
Haig D (2002) Genomic imprinting and kinship. Rutgers University Press, New Brunswick
Charlesworth B, Charlesworth D (2000) The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci 355(1403):1563–1572
Charlesworth B (1978) Model for evolution of Y chromosomes and dosage compensation. Proc Natl Acad Sci USA 75(11):5618–5622
Charlesworth B (1996) The evolution of chromosomal sex determination and dosage compensation. Curr Biol 6(2):149–162
Acknowledgments
We thank Fangqin Lin for providing us with the exact X:AA and X:XX median values from [34]. GABM is supported by Agence Nationale de la Recherche (Grant ref. ANR-12-BSV7-0002). GABM thanks Instituto Gulbenkian de Ciência for hosting him during several periods strongly overlapping with the writing of this article.
Author information
Authors and Affiliations
Corresponding author
Appendices
Box 1: testing Ohno’s hypothesis with expression data
Ohno’s hypothesis has been tested by comparing the expression of the X chromosome to that of the autosomes taken together, the X:AA ratio.
Microarray versus RNAseq data
Initially, microarray data have been used for this test [22–26]. Microarray may give less precise estimates of expression levels [27]. Moreover, microarray data have to be filtered prior to analysis. The procedures for data filtering rely on arbitrary thresholds, which applied similarly to the X and autosomes remove many lowly expressed X-linked genes and generate an artifactual X:AA of 1 [68]. RNAseq data are supposed to give more precise estimates of expression levels. However, there is also some noise in RNAseq data due to unspecific mapping of RNAseq reads onto the genome, and how this noise is removed can also affect the results [69]. Removing this noise is at the heart of the controversy between the different studies using RNAseq [27–30, 35]. As the threshold for considering a given expression level different from 0 increases, the X:AA ratio increases and reaches a plateau at 1 [28]. It is clear, however, that when using conservative thresholds, the number of X-linked genes analyzed becomes small, and one cannot conclude from this about a “global” X hyperactivation [32].
X:AA, X:XX, and other expression ratios
Using X:AA expression relies on the assumption that expression were similar between the proto-sex chromosomes and the autosomes (XX:AA = 1). Using the present-day and ancestral expression of the X chromosome, the X:XX ratio, is thus a more direct way to test for Ohno’s hypothesis. Computing the X:XX ratio in mammals has implied finding an outgroup where the 1-to-1 orthologs of the X-linked genes are autosomal, namely birds [33]. This guarantees that only genes that were originally on the sex chromosomes before they diverged are analyzed, which is what should be done as dosage compensation is expected for these genes only. The new genes that evolved (e.g. through intra-X duplication or translocation to the X) after X and Y stopped recombining and diverged should not be included in studies on dosage compensation, are correctly excluded of the X:XX analysis but not in the X:AA ones. However, finding 1-to-1 orthologs between distantly related species may be difficult and result in a small number of genes being analyzed. Moreover, all these chromosome-wide comparisons may be problematic as different selective forces (dosage compensation, sexual selection) may affect expression levels [43, 70]. A more precise way of testing Ohno’s hypothesis is to study X-linked and autosomal genes that are expected to interact in some ways and for which equal dosage may be required. Considering genes from the same network is one possibility [33], and considering genes belonging to protein complexes is another [32].
Box 2: The parental antagonism model of X chromosome inactivation
The parental antagonism model (PAM) for the evolution of XCI was proposed by Haig [49, 50]. It is embedded within the general evolutionary theory of parental investment in offspring [71] and closely related to the kinship theory of genomic imprinting [49, 72]. The argument can be presented in a number of steps.
Step 0 A prerequisite for PAM to work is that offspring are provisioned with an adjustable amount of resources from their mother following fertilization. This is indeed the case in therians where resources are provided through the placenta during embryonic development.
Step 1 At the core of PAM is the expectation that there will be an evolutionary conflict between maternally and paternally derived genes within a developing organism with respect to the amount of resources provided by the mother of that individual. Both genes derived from the mother and from the father will be selected to induce the mother to provide resources. However, the optimal amount of resources provided may be greater for paternally than for maternally derived genes. This is because when females mate with multiple males during their lifetimes, paternal interests will be limited to the current offspring whereas maternal interests extend to all future offspring that a mother will have.
Step 2 The X chromosome is two-thirds of the time inherited from the mother but only one-third of the time from the father (simply because females have two Xs and males just one). As a consequence, genes on the X chromosome are expected to reflect maternal interests more than paternal ones. In particular, it is expected that genes coding for embryonic growth inhibitors will accumulate on the X chromosome, whereas growth enhancers will be scarce [51].
Step 3 As an evolutionary response to this accumulation of growth inhibitor genes on the X chromosome, there will be selection on paternally inherited genes on the X chromosome to inactivate these genes in embryos, thereby increasing embryo growth. This inactivation may then also spread to other genes on the paternally derived X, either for mechanistic reasons or for dosage compensation. The resulting state of inactivation of the paternally derived X (pXCI) is found in marsupials.
Step 4 pXCI entails that an organism becomes functionally haploid, so that recessive deleterious mutations on the maternally derived X chromosome will be expressed and reduce fitness. This may create selection pressure for random XCI (rXCI), alleviating this burden because half of the cells will then express the functional gene copy [50]. This transition from pXCI to rXCI does not involve parental conflict because the choice of which X chromosome is inactivated does not affect gene dosage.
Step 5 Nevertheless, parental conflict over which of the X chromosomes is inactivated may persist or re-emerge. This is because there may still be imprinted growth inhibitor genes on the X chromosome that are silenced when paternally inherited, so that the maternally derived X chromosome will be under selection to remain the active X. As a consequence, pXCI can re-evolve from rXCI, which may explain pXCI in mouse trophoblast tissues.
Rights and permissions
About this article
Cite this article
Pessia, E., Engelstädter, J. & Marais, G.A.B. The evolution of X chromosome inactivation in mammals: the demise of Ohno’s hypothesis?. Cell. Mol. Life Sci. 71, 1383–1394 (2014). https://doi.org/10.1007/s00018-013-1499-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-013-1499-6