Abstract
Introduction
Circulating IL-6 levels and at least one polymorphic form of IL6 gene (IL6 -174 G/C, rs1800795) have been shown to be independently associated with coronary artery disease (CAD) by several investigators. Despite more than 12 published meta-analyses on this subject, association of -174 G/C with CAD, especially amongst distinct ancestral population groups remain unclear. We, therefore, conducted a systematic review and an updated meta-analysis to comprehensively ascertain the association of IL6 -174 G/C with CAD and circulating IL-6 levels.
Materials and methods
Relevant case–control/cohort studies investigating association of -174 G/C with CAD and circulating IL-6 levels were identified following a comprehensive online search. Association status for CAD was determined for the pooled sample, as well as separately for major ancestral subgroups. Association status for circulating IL-6 levels was assessed for the pooled sample, as well as separately for CAD cases and CAD free controls. Study-level odds ratios (OR) and 95% confidence intervals (CI) were pooled using random/fixed-effects model.
Results
Quantitative synthesis for the CAD endpoint was performed using 55 separate qualifying studies with a collective sample size of 51,213 (19,160 cases/32,053 controls). Pooled association of -174 G/C with CAD was found to be statistically significant through dominant (OR 1.15; 95% CI 1.05–1.25, p = 0.002) as well as allelic genetic model comparisons (OR 1.13, 95% CI 1.06–1.21, p = 0.0003). This effect was largely driven by Asian and Asian Indian ancestral subgroups, which also showed significant association with CAD in both genetic model comparisons (OR range 1.29–1.53, p value range ≤ 0.02). Other ancestral subgroups failed to show any meaningful association.
Circulating IL-6 levels were found to be significantly higher amongst the ‘C’ allele carriers in the pooled sample (Standard mean difference, SMD 0.11, 95% CI 0.01–0.22 pg/ml, p = 0.009) as well as in the CAD free control subgroup (SMD 0.10, 95% CI 0.02–0.17 pg/ml, p = 0.009), though not in the CAD case subgroup (SMD 0.17, 95% CI = − 0.02 to 0.37, p = 0.12).
Conclusions
The present systematic review and meta-analysis demonstrate an overall association between IL6 -174 G/C polymorphism and CAD, which seems to be mainly driven by Asian and Asian Indian ancestral subgroups. Upregulation of plasma IL-6 levels in the ‘C’ allele carriers seems to be at least partly responsible for this observed association. This warrants further investigations with large, structured case–control studies especially amongst Asian and Asian Indian ancestral groups.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
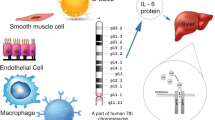
Interleukin 6 (IL-6) is a circulating bioactive peptide of 23.7 kDa and acts as both a pro-inflammatory cytokine and an anti-inflammatory myokine. This endogenous pyrogen primarily originates from mononuclear phagocytes but also, in part, from fibroblasts, T and B lymphocytes and vascular endothelial cells [1, 2]. It functions in inflammation and maturation of B cells [3], and is encoded by IL6 gene (located at chromosome 7p21–14) which is known to have several polymorphic variants [4]. Circulating IL-6 levels and at least one polymorphic form of IL6 gene have been reported to be independently associated with coronary artery disease (CAD), at least amongst Caucasians [5, 6].
G to C substitutions at -174 bp (-174 G/C; rs1800795) and -572 bp (-572 G/C, rs1800796) in the promoter region of IL6 gene are the most important single nucleotide polymorphisms (SNPs) with respect to CAD [5,6,7]. These two SNPs have been reported to be functional, seem to have a co-operative influence over each other and are capable of altering circulating IL-6 levels through complex interactions depending on the haplotype [4, 8]. There have been at least 12 published meta-analyses investigating the association of common IL6 SNPs with CAD, myocardial infarction (MI; or surrogates), and related clinical presentations. [5,6,7, 9,10,11,12,13,14,15,16,17] However, flawed ethnic stratifications in these meta-analyses yielded inconsistent results [5,6,7, 9,10,11,12,13,14,15,16,17]. Against this background, our objectives in this systematic review and meta-analysis were to: (i) ascertain the overall association of IL6 -174 G/C polymorphism with CAD, as well as separately amongst different ancestral populations, (ii) investigate the association of this polymorphic form with circulating IL-6 levels.
Material and methods
Relevant guidelines in the HuGE Review Handbook, version 1.0 [18] as well as the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement [19] were strictly adhered to while undertaking the present systematic review and meta-analysis.
Search strategy and study selection criteria
We systematically searched the databases of the US National Institutes of Health (PubMed), EMBASE, MEDLINE, Scopus and Web of Knowledge for relevant articles published online until May 2021. Specific search headings as well as open text fields were used for the online publication search. Reference lists of relevant published meta-analyses were also scanned for identifying additional articles. Combination of broad search headings such as ‘interleukin 6’ OR ‘IL6’ OR rs1800795 (dbSNP ID’s or rs number) paired with ‘coronary artery disease’ OR ‘CAD’ OR ‘myocardial infarction’ OR ‘MI’ OR ‘acute coronary syndrome’ OR ‘ACS’ AND ‘polymorphism’ OR ‘mutation’ OR ‘single nucleotide polymorphism’ OR ‘SNP’ were used for online search. Our search was limited to publications in the English language and restricted to articles relating to humans.
Hierarchical model for study selection was used: initially the study title was assessed for relevance, followed by the abstract and finally, the full text. To qualify for inclusion, the relevant study had to be either a case–control study or cohort study with a well-documented CAD case group (diagnosed CAD, MI, ACS, unstable/stable angina pectoris) compared against a CAD free control group. To be included, all studies had to satisfy the following criteria: (1) original, published in a peer-reviewed journal, and available online, (2) case–control or cohort design, (3) providing complete genotypic data crucial for calculation of odds ratios (OR), confidence intervals (CIs) and p-values, (4) CAD diagnosis amongst cases had to be based on angiographic or electrocardiographic assessment whilst controls had to be free of any history or evidence of CAD, (5) published in the English language with online accessibility, and (6) genotype frequencies amongst controls satisfying Hardy–Weinberg equilibrium (HWE). We assessed departure from HWE amongst controls in each study using the goodness-of-fit × 2 test. Incidences of non-conformation to HWE approximation (p < 0.05) resulted in the exclusion of the study. Conference abstracts and case reports/studies not providing adequate information were also excluded. Publications lacking enough data to generate both dominant and allelic genetic models were identified and their corresponding authors were formally requested for supplying missing data via three periodic emails, spaced 1 week apart. We included the study after receiving the complete data. In case all efforts to retrieve the missing data failed, we included studies where enough data was available to construct at least one genetic model. If relevant data was not made available even after three consecutive requests and published data was not sufficient to construct even one genetic model, the study in question was excluded. Further study selection to ascertain the association of -174 G/C polymorphism with circulating IL-6 levels was done from the already searched publications.
Data collection and quality assessment
Raw data were transcribed from selected publications on Microsoft-Excel worksheets where further calculations were performed. Studies qualifying for testing association of -174 G/C with CAD were stratified into ancestral subgroups such as European ancestry, Middle Eastern ancestry, Asian ancestry, Asian Indian ancestry, African ancestry and Mixed ancestry. Studies were categorized based on the ancestral background of the majority of the studied population. Studies were stratified into ‘CAD cases’ and ‘CAD free control’ subgroups while testing for an association of -174 G/C polymorphism with circulating IL-6 levels.
Quality assessment of the included studies was performed using Newcastle–Ottawa scale (NOS) (http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp). The NOS is a star-based rating system where a study with a full score can earn 9 stars. A NOS rating of 5–9 stars was indicative of a good quality study, while a score of 0–4 stars indicated a poor quality study [20]. The NOS rating tool involves evaluation of (a) selection methods of study participants, (b) comparability amongst cases and control groups, and (c) exposure and outcome. Included studies were independently assessed for quality by both authors; disagreements were then resolved by consensus.
Environmental factors have been known to have a profound impact on the association profiles of genetic polymorphisms. Since none of our included studies provided raw calculable data on environmental factors, we were unable to test the impact of this relationship.
Statistical techniques
Calculations were carried out using windows based RevMan version 5.3.5 (The Cochrane Collaboration, 2014) and SPSS version 25 (IBM® corporation).
Summary effect measures
Odds ratios (ORs) were calculated using bivariate, random (DerSimonian–Laird method) [21] or fixed-effect model (Mantel–Haenszel method) [22]. Summary ORs and their 95% confidence intervals (CIs) were calculated separately for dominant and allelic genetic models. Analytic models (random or fixed) were chosen based on observed heterogeneity within the group/subgroup. The calculated OR and 95% CI for each study revealed the level of association (if any). The pooled OR was estimated from individual study ORs employing a Z test. The summary effect measure for the association of -174 G/C with circulating IL-6 levels was pooled standard mean difference (SMD) with its 95%CI (in pg/ml). SMDs were estimated for each study after which a Z test was employed to ascertain a pooled SMD. For both summary effect measures, a pooled p value of < 0.05 indicated statistical significance and the corresponding Z value indicated the level of association.
Heterogeneity assessment
Existence of heterogeneity was tested using a Q test. Resulting Higgins I2 statistics (I2) and Cochrane’s Q statistics (PQ) for each study group/subgroup indicated inherent heterogeneity. A heterogeneous group/subgroup was assumed to be with a resultant PQ cut-off < 0.01 [23]. The I2 value cut offs of 25%, 50% and 75% indicated low, moderate and high heterogeneity, respectively [24]. Random effects for calculation of summary effect measures were used if the group/subgroup yielded a PQ value of ≤ 0.01 coupled with an I2 value of ≥ 50%. Conversely, fixed effect was used for summary effect measure estimation if the group/subgroup yielded a PQ value of > 0.01 coupled with an I2 value of < 50%. Subgroup differences were also assessed assuming similar PQ and I2 cut-offs.
Detection of publication bias
We employed two of the most accepted statistical tools for publication bias detection in the present meta-analysis. Publication bias in each group of ≥ 3 studies was visually detected using Begg’s funnel plot [25], while the statistical estimates for each group/subgroup were calculated using Egger’s test.[26] An Egger’s p value of < 0.05 was considered statistically significant and indicated the possible existence of publication bias in the group/subgroup in question.
Sensitivity analysis
Sensitivity analysis was performed separately in each study group/subgroup (with ≥ 5 studies). We repeated the analysis after the omission of one study after another in each qualifying group/subgroup. This exercise was performed to see if the results in any group/subgroup altered substantially, i.e. a change from non-association to a significant association or the other way around. Absence of such alteration in results indicates the robustness of the meta-analysis in question.
Results
Screening of 394 records led to the identification of 47 relevant articles. A total of 55 different studies (extracted from 47 articles) were included to test the association of -174 G/C with CAD [27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73]. The study selection process is explained in detail in Fig. 1. Table 1 contains complete details of all included studies for the CAD endpoint, while Table 2 lists studies included for circulating IL-6 levels endpoint. Sample assessed and inherent heterogeneity of studied groups and subgroups included for CAD endpoint are shown in Supplementary Table 1. Meta-analysis results obtained for CAD endpoint are summarized in Supplementary Table 2.
Role of -174 G/C polymorphism in CAD
A total of 55 case–control/cohort genetic association studies on -174 G/C, with a total sample of 51,213 (19,160 cases/32,053 controls) were analyzed [27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73]. The pooled group showed significant heterogeneity as the included studies belonged to 6 distinct ancestral subgroups. (Supplementary Table 1) Our pooled results via both genetic models were obtained using random effects. Pooled comparisons using dominant (CC + GC vs. GG) and allelic genetic models (allele C vs. allele G) suggested statistically significant association with CAD [number of studies (nst) = 52, OR 1.15, 95% CI 1.05–1.25, Z = 3.03 and p = 0.002 and nst = 53, OR 1.13, 95% CI 1.06–1.21, Z = 3.65 and p = 0.0003, respectively]. (Fig. 2, Panel A and Fig. 3, Panel A, respectively, for dominant and allelic model, Supplementary Figs. 1 and 2, respectively, for dominant and allelic model and Supplementary Table 2).
Meta-analysis results and publication bias assessment employing dominant genetic model comparisons (CC + GC versus GG) for CAD endpoint. Panel A Condensed Forest plot depicting associations of IL6 -174 G/C polymorphism with CAD. R Effect sizes for “Pooled” as well as for European and Asian Indian ancestral subgroups displaying high level of heterogeneity were estimated using random effects for analysis. Effect sizes for Middle Eastern, Asian, African and Mixed ancestral subgroups were estimated using fixed effects. Effect sized are displayed as Odds Ratio (95% Confidence Interval). *Statistically significant p value of < 0.05. Panel B Publication bias assessment using Begg’s funnel plot with Egger’s estimates amongst the group of studies investigating the role IL6 -174 G/C polymorphism in CAD. Each point in this figure represents the odds ratio (OR) obtained for a study plotted against its standard error (SE). Different indicators have been used for studies belonging to each ancestral subgroup. *Statistically significant p value of < 0.05
Meta-analysis results and publication bias assessment employing allelic genetic model comparisons (Allele C versus Allele G) for CAD endpoint. Panel A Condensed Forest plot depicting associations of IL6 -174 G/C polymorphism with CAD. R Effect sizes for “Pooled” as well as for European and Asian Indian ancestral subgroups displaying a high level of heterogeneity were estimated using random effects for analysis. Effect sizes for Middle Eastern, Asian, African and Mixed ancestral subgroups were estimated using fixed effects. Effect sized are displayed as Odds Ratio (95% Confidence Interval). *Statistically significant p value of < 0.05. Panel B Publication bias assessment using Begg’s funnel plot with Egger’s estimates amongst the group of studies investigating the role IL6 -174 G/C polymorphism in CAD. Each point in this figure represents the odds ratio (OR) obtained for a study plotted against its standard error (SE). Different indicators have been used for studies belonging to each ancestral subgroup. *Statistically significant p value of < 0.05
Similarly, summary effect measures for all ancestral subgroups, respectively, were also obtained using appropriate effects for analysis. Results obtained for Asian ancestry [54,55,56,57,58,59, 72] and Asian Indian ancestry [60,61,62,63,64,65,66,67,68, 70, 71] subgroups displayed significant positive association via both genetic models (nst = 7, OR range 1.46–1.53, Z value range 6.04–7.03, p value range = < 0.0001 to < 0.00001 and nst = 12, OR range 1.29–1.31, Z value range = 2.28–2.95, p value range = 0.02–0.003, respectively). On the other hand, Mixed ancestry subgroup [73] showed a negative association with CAD (nst = 1, OR range 0.42–0.55, Z value range = 2.71–3.50, p value range = 0.007–0.0005). Other ancestral subgroups such as European ancestry [27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48, 69], Middle Eastern ancestry [49,50,51,52,53] and African ancestry [45] did not seem to be associated with CAD (p value range = 0.11–0.47). (Fig. 2, Panel A and Fig. 3, Panel A, respectively, for dominant and allelic model, Supplementary Figs. 1 and 2, respectively, for dominant and allelic model and Supplementary Table 2) Understandably, substantial subgroup differences were observed (I2 range = 82.5–84.1% and PQ range = <0.0001 to <0.00001) (Supplementary Table 2).
Association of -174 G/C polymorphism with circulating IL-6 levels
IL-6 levels amongst ‘C’ allele carriers were compared against the rest in the overall sample and separately amongst CAD cases (nst = 9; 3418 subjects) [29, 31, 33, 35, 46, 65, 66, 74] and CAD free controls (nst = 6; 3389 subjects) [29, 31, 33, 46, 66]. ‘C’ allele carriers in the overall sample were associated with significantly higher plasma levels of IL-6 (SMD 0.11, 95% CI 0.01–0.22 pg/ml, Z value = 2.14, p = 0.03). While CAD case subgroup did not yield significant association (p = 0.12), IL-6 levels were found to be significantly higher amongst the ‘C’ allele carriers in the CAD free control subgroup (SMD 0.10, 95% CI 0.02–0.17 pg/ml, Z value = 2.62, p = 0.009) as shown in Fig. 4, Panel A and Supplementary Fig. 3. No evidence of publication bias was seen in pooled group as well as in the subgroups, which validated the derived associations (Egger’s p value range = 0.663–0.918) (Fig. 4, Panel B).
Meta-analysis results depicting differences in circulating IL-6 levels amongst ‘C’ allele carriers versus GG homozygotes as well as publication bias assessment results in the included groups/subgroups. Panel A Comparison of IL-6 levels between ‘C’ allele carriers as compared to the rest (CC+GC vs. GG) separately amongst CAD cases and CAD free controls. RStandard mean difference for “Pooled” as well as CAD case subgroup were estimated using random effects owing to high levels of inherent heterogeneity. Standard mean difference for CAD free control subgroup which displayed low levels of inherent heterogeneity were estimated using fixed effects. Standard mean difference and its 95% Confidence Interval is depicted in the bar charts. *Statistically significant p value of < 0.05. Panel B Begg’s funnel plot with Egger’s estimates was obtained for comparison of circulating IL-6 levels between ‘C’ allele carries as compared to the rest (CC + GC vs. GG). *Statistically significant p value of < 0.05. Each point in each figure represents the standard mean difference (SMD, in pg/ml) obtained for a study plotted against its standard error (SE). Different indicators have been used for studies belonging to ‘CAD cases’ and ‘CAD free controls’ subgroups
Publication bias assessment and sensitivity analysis
Each group or subgroup with ≥ 3 included studies was assessed for existing publication bias using Begg’s funnel plot test [25] and Egger’s test [26]. Begg’s funnel plots and Egger’s p values for each qualifying group/subgroup constructed for -174 G/C for CAD endpoint are displayed in Fig. 2, Panel B and Fig. 3, Panel B (respectively, for dominant and allelic model), while for circulating IL-6 levels endpoint in Fig. 4, Panel B. Each point in these plots represents the OR or SMD obtained for an included study plotted against its standard error (SE). Different indicators have been used for studies belonging to different ancestral subgroups/CAD cases or CAD free control subgroups. All these points seem to be generally contained within the inverted cone, indicating limited existence publication bias. Egger’s p values seem to reach statistical significance for most of the ancestral groups and subgroups which could have been a direct result of inherent heterogeneity. This indicates that the use of ancestral stratification was also not sufficient to tone down the possible existence of bias. On the other hand, we found no evidence of publication bias in subgroups constructed for circulating IL-6 endpoint.
Sensitivity analysis was performed in each study group/subgroup with ≥ 5 included studies. Studies were excluded one after another in these groups/subgroups and the analysis was repeated after each omission. We observed no instance of significant alteration from the original results, i.e. from lack of association to significant association or the other way around for both endpoints, which is an indicator of the robustness of the meta-analysis in question (Data not shown).
Discussion
We present the most comprehensive and structured meta-analysis on the association between IL6 -174 G/C polymorphism with CAD as well as circulating IL-6 levels. The main findings were: (i) pooled results indicated a significant association of -174 G/C polymorphism with CAD; however, the effect was driven by studies with participants belonging to Asian and Asian Indian ancestries; (ii) other major ancestries, including European and Middle Eastern displayed no evidence of such association; (iii) ‘C allele’ carriers, at least amongst CAD free controls seem to have significantly higher levels of circulating IL-6, which in part explains the association of this SNP with CAD.
Results obtained in our meta-analysis for the CAD endpoint are much more robust than the recent one on this subject [17], which incidentally also lacks ancestral stratification needed to identify drivers of seen association. The latest meta-analysis which is comparable to ours’ was from Hou and coworkers published in 2015 [6], Our results for -174 G/C polymorphism with CAD represents a complete shift from their results. First, our pooled results displayed overwhelmingly strong associations with CAD (p ≤ 0.0005, for both genetic models), in contrast to a milder level of associations (p = 0.01 in both models) reported by Hou et al. [6] Second, we tried to correctly stratify different ancestral populations into appropriate subgroups thus revealing a clear picture, which is in contrast to Hou et al. [6], who clubbed Europeans along with Indian, Turkish, Tunisian and Pakistani populations in their ‘Caucasian’ subgroup.
At least 4 promoter polymorphisms of the IL6 gene at positions 597, 572, 373 along with 174 bp, have been known to influence IL6 transcription through complex interactions determined by the haplotype [4]. We hypothesize that ‘C’ allele carriers in -174 G/C, through a variety of mechanisms, are more likely to have upregulated transcription and translation of IL6 gene; are, therefore, associated with higher plasma concentrations of circulating IL-6, thus making them more susceptible to the development of atherosclerotic disease. We tested this hypothesis and found that while possibly the influence of concomitant medications [75], prevented the CAD case subgroup to yield significant association (p = 0.12), our CAD free control subgroup showed a clear association of ‘C’ allele carriers with elevated circulating IL-6 levels (p = 0.009).
This difference has also been observed locally at the transcriptional level where IL6 mRNA expression is 10–40 fold higher in atherosclerotic as compared to healthy arteries [76]. IL-6 not only has a direct association with CAD; it also indirectly contributes to the development of atherosclerotic disease in several ways. Circulating IL-6 has been known to regulate fibrinogen—an acute-phase protein which is recognized as an important risk factor for atherosclerotic and thrombotic diseases [76]. It has also been reported to stimulate the differentiation of monocytes to macrophages, which contributes towards the growth of atherosclerotic plaques [77]. The effect of individual IL6 gene SNPs on the regulation of plasma IL-6 levels have also been investigated before [4]. Since, at least 4 adjacent IL6 polymorphic sites (-174 G/C, -373 A/T, -572 G/C and -597 G/A) have complex interactions between each of their transcriptional machineries [4], it this not easy to determine the effects of a single variant. IL6 promoter haplotypes have been reported to be better predictors of transcription levels of IL6 gene [4]. Investigating the synergistic effect of these possible haplotypes on CAD, MI or circulating IL-6 levels was not possible due to the lack of relevant published haplotypic data.
Limitations
Meta-analyses on genetic association studies tend to have significant limitations. First, for some ancestral subgroups—only a few published reports with moderate sample sizes were available; their meta-analysis results should thus be interpreted with caution. More studies from these ancestral groups are warranted to establish these derived associations. Second, the fact that meta-analyses of association studies cannot inspect interference of linkage disequilibrium, it constitutes as a major limitation. Third, the presence of selection bias in individual included studies and the presence of publication bias in a meta-analysis of non-randomized, genetic association studies easily qualify to be the most important limitation. Several statistical tools are available to test publication bias, although none are perfect, are easily influenced by heterogeneity, and in our case two of them were used which gave inconsistent results. This fact illustrates that the role of existing publication bias cannot be completely ruled out. We cannot be sure whether to trust our funnel plots where most of the studies were contained within the inverted cone, signaling a lack of publication bias or the results of the Egger’s test where significant p values were seen for both genetic model comparisons in most of the analyzed groups/subgroups.
Conclusions
Significant association of IL6-174 G/C variant with CAD was observed in the pooled results of our present meta-analysis, largely driven by studies belonging to Asian and Asian Indian ancestral subgroups. Upregulation of plasma IL-6 levels in the ‘C’ allele carriers seem to be at least partly responsible for this seen association. Further investigations are warranted with large structured case–control studies especially amongst populations belonging to Asian and Asian Indian ancestry.
References
Talaat RM, Abdel-Aziz AM, El-Maadawy EA, Abdel-Bary N. CD38 and interleukin 6 gene polymorphism in Egyptians with diffuse large B-cell lymphoma (DLBCL). Immunol Investig. 2015;44(3):265–78. https://doi.org/10.3109/08820139.2014.989328.
Zhang K, Zhang L, Zhou J, Hao Z, Fan S, Yang C, et al. Association between interleukin-6 polymorphisms and urinary system cancer risk: evidence from a meta-analysis. OncoTargets Therapy. 2016;9:567–77. https://doi.org/10.2147/ott.s94348.
Jia W, Fei GH, Hu JG, Hu XW. A study on the effect of IL-6 gene polymorphism on the prognosis of non-small-cell lung cancer. OncoTargets Therapy. 2015;8:2699–704. https://doi.org/10.2147/ott.s84636.
Terry CF, Loukaci V, Green FR. Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem. 2000;275(24):18138–44. https://doi.org/10.1074/jbc.M000379200.
Niu W, Liu Y, Qi Y, Wu Z, Zhu D, Jin W. Association of interleukin-6 circulating levels with coronary artery disease: a meta-analysis implementing Mendelian randomization approach. Int J Cardiol. 2012;157(2):243–52. https://doi.org/10.1016/j.ijcard.2011.12.098.
Hou H, Wang C, Sun F, Zhao L, Dun A, Sun Z. Association of interleukin-6 gene polymorphism with coronary artery disease: an updated systematic review and cumulative meta-analysis. Inflamm Res. 2015;64(9):707–20. https://doi.org/10.1007/s00011-015-0850-9.
Liu SL, Yin YW, Sun QQ, Hu AM, Zhang SJ. Genetic polymorphisms of interleukin-6 gene and susceptibility to coronary artery disease in Chinese population: Evidence based on 4582 subjects. Human Immunol. 2015;76(7):505–10. https://doi.org/10.1016/j.humimm.2015.06.004.
Rivera-Chavez FA, Peters-Hybki DL, Barber RC, O’Keefe GE. Interleukin-6 promoter haplotypes and interleukin-6 cytokine responses. Shock (Augusta, Ga). 2003;20(3):218–23.
Li YY, Zhou CW, Xu J, Qian Y, Wang XM. Interleukin-6 C-572G gene polymorphism and coronary artery disease in Asian: a meta-analysis of 2511 subjects. Int J Clin Exp Med. 2015;8(6):8995–9003.
Ma Y, Tang RK, Yang X, Peng GG, Liu Y, Wang XM, et al. Lack of an association between interleukin-6 gene promoter polymorphisms (-174G/C, -572G/C) and ischemic heart disease and/or ischemic stroke: a meta-analysis. Human Immunol. 2011;72(8):641–51. https://doi.org/10.1016/j.humimm.2011.03.019.
Ren H, Zhang Y, Yao Y, Guo T, Wang H, Mei H, et al. Association between the interleukin-6 genetic polymorphism 174 G/C and thrombosis disorder risk: meta-analysis of 10,549 cases and 19,316 controls. Medicine. 2016;95(27):e4030. https://doi.org/10.1097/md.0000000000004030.
Yang Y, Zhang F, Skrip L, Lei H, Wang Y, Hu D, et al. IL-6 gene polymorphisms and CAD risk: a meta-analysis. Mol Biol Rep. 2013;40(3):2589–98. https://doi.org/10.1007/s11033-012-2345-x.
Yin YW, Hu AM, Sun QQ, Liu HL, Wang Q, Zeng YH, et al. Association between interleukin-6 gene -174 G/C polymorphism and the risk of coronary heart disease: a meta-analysis of 20 studies including 9619 cases and 10,919 controls. Gene. 2012;503(1):25–30. https://doi.org/10.1016/j.gene.2012.04.075.
Yin YW, Li JC, Zhang M, Wang JZ, Li BH, Liu Y, et al. Influence of interleukin-6 gene -174G>C polymorphism on development of atherosclerosis: a meta-analysis of 50 studies involving 33,514 subjects. Gene. 2013;529(1):94–103. https://doi.org/10.1016/j.gene.2013.07.074.
Zheng GH, Chen HY, Xiong SQ. Polymorphisms of -174G>C and -572G>C in the interleukin 6 (IL-6) gene and coronary heart disease risk: a meta-analysis of 27 research studies. PLoS ONE. 2012;7(4):e34839. https://doi.org/10.1371/journal.pone.0034839.
Lu S, Wang Y, Wang Y, Hu J, Di W, Liu S, et al. The IL-6 rs1800795 and rs1800796 polymorphisms are associated with coronary artery disease risk. J Cell Mol Med. 2020;24(11):6191–207. https://doi.org/10.1111/jcmm.15246.
Tabaei S, Motallebnezhad M, Tabaee SS. Systematic review and meta-analysis of association of polymorphisms in inflammatory cytokine genes with coronary artery disease. Inflamm Res. 2020;69(10):1001–13. https://doi.org/10.1007/s00011-020-01385-3.
Little J, Higgins J, Bray M, Ioannidis J, Khoury M, Manolio T, et al. The HuGENet™ HuGE review handbook, version 1.0. Ottawa, Ontario, Canada: HuGENet Canada Coordinating Centre. 2006.
Panic N, Leoncini E, de Belvis G, Ricciardi W, Boccia S. Evaluation of the endorsement of the preferred reporting items for systematic reviews and meta-analysis (PRISMA) statement on the quality of published systematic review and meta-analyses. PLoS ONE. 2013;8(12):e83138. https://doi.org/10.1371/journal.pone.0083138.
Rai H, Sengupta S, Sinha N, Kumar S, Sharma AK, Agrawal S. Interleukin-1 gene cluster polymorphisms and their association with coronary artery disease: separate evidences from the largest case-control study amongst north Indians and an updated meta-analysis. PLoS ONE. 2016. https://doi.org/10.1371/journal.pone.0153480.
DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–88.
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–48.
Rai H, Parveen F, Kumar S, Kapoor A, Sinha N. Association of endothelial nitric oxide synthase gene polymorphisms with coronary artery disease: an updated meta-analysis and systematic review. PLoS ONE. 2014;9(11):e113363. https://doi.org/10.1371/journal.pone.0113363.
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical research ed). 2003;327(7414):557–60. https://doi.org/10.1136/bmj.327.7414.557.
Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–101.
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical research ed). 1997;315(7109):629–34.
Humphries SE, Luong LA, Ogg MS, Hawe E, Miller GJ. The interleukin-6 -174 G/C promoter polymorphism is associated with risk of coronary heart disease and systolic blood pressure in healthy men. Eur Heart J. 2001;22(24):2243–52. https://doi.org/10.1053/euhj.2001.2678.
Georges JL, Loukaci V, Poirier O, Evans A, Luc G, Arveiler D, et al. Interleukin-6 gene polymorphisms and susceptibility to myocardial infarction: the ECTIM study. Etude Cas-Temoin de l’Infarctus du Myocarde. J Mol Med (Berlin, Germany). 2001;79(5–6):300–5.
Basso F, Lowe GD, Rumley A, McMahon AD, Humphries SE. Interleukin-6 -174G>C polymorphism and risk of coronary heart disease in West of Scotland coronary prevention study (WOSCOPS). Arterioscler Thromb Vasc Biol. 2002;22(4):599–604.
Nauck M, Winkelmann BR, Hoffmann MM, Bohm BO, Wieland H, Marz W. The interleukin-6 G(-174)C promoter polymorphism in the LURIC cohort: no association with plasma interleukin-6, coronary artery disease, and myocardial infarction. J Mol Med (Berlin, Germany). 2002;80(8):507–13. https://doi.org/10.1007/s00109-002-0354-2.
Bennet AM, Prince JA, Fei GZ, Lyrenas L, Huang Y, Wiman B, et al. Interleukin-6 serum levels and genotypes influence the risk for myocardial infarction. Atherosclerosis. 2003;171(2):359–67.
Georges JL, Rupprecht HJ, Blankenberg S, Poirier O, Bickel C, Hafner G, et al. Impact of pathogen burden in patients with coronary artery disease in relation to systemic inflammation and variation in genes encoding cytokines. Am J Cardiol. 2003;92(5):515–21.
Kelberman D, Hawe E, Luong LA, Mohamed-Ali V, Lundman P, Tornvall P, et al. Effect of Interleukin-6 promoter polymorphisms in survivors of myocardial infarction and matched controls in the North and South of Europe. The HIFMECH Study. Thrombos Haemostas. 2004;92(5):1122–8. https://doi.org/10.1160/th04-04-0202.
Licastro F, Chiappelli M, Caldarera CM, Tampieri C, Nanni S, Gallina M, et al. The concomitant presence of polymorphic alleles of interleukin-1beta, interleukin-6 and apolipoprotein E is associated with an increased risk of myocardial infarction in elderly men. Results from a pilot study. Mech Ageing Dev. 2004;125(8):575–9. https://doi.org/10.1016/j.mad.2004.06.008.
Lieb W, Pavlik R, Erdmann J, Mayer B, Holmer SR, Fischer M, et al. No association of interleukin-6 gene polymorphism (-174 G/C) with myocardial infarction or traditional cardiovascular risk factors. Int J Cardiol. 2004;97(2):205–12. https://doi.org/10.1016/j.ijcard.2003.07.038.
Chiappelli M, Tampieri C, Tumini E, Porcellini E, Caldarera CM, Nanni S, et al. Interleukin-6 gene polymorphism is an age-dependent risk factor for myocardial infarction in men. Int J Immunogenet. 2005;32(6):349–53. https://doi.org/10.1111/j.1744-313X.2005.00537.x.
Densem CG, Ray M, Hutchinson IV, Yonan N, Brooks NH. Interleukin-6 polymorphism: a genetic risk factor for cardiac transplant related coronary vasculopathy? J Heart Lung Transplant. 2005;24(5):559–65. https://doi.org/10.1016/j.healun.2004.03.016.
Rosner SA, Ridker PM, Zee RY, Cook NR. Interaction between inflammation-related gene polymorphisms and cigarette smoking on the risk of myocardial infarction in the Physician’s Health Study. Hum Genet. 2005;118(2):287–94. https://doi.org/10.1007/s00439-005-0052-6.
Seifart C, Dempfle A, Plagens A, Seifart U, Clostermann U, Muller B, et al. TNF-alpha-, TNF-beta-, IL-6-, and IL-10-promoter polymorphisms in patients with chronic obstructive pulmonary disease. Tissue Antigens. 2005;65(1):93–100. https://doi.org/10.1111/j.1399-0039.2005.00343.x.
Sie MP, Sayed-Tabatabaei FA, Oei HH, Uitterlinden AG, Pols HA, Hofman A, et al. Interleukin 6–174 g/c promoter polymorphism and risk of coronary heart disease: results from the rotterdam study and a meta-analysis. Arterioscler Thromb Vasc Biol. 2006;26(1):212–7. https://doi.org/10.1161/01.atv.0000194099.65024.17.
Sarecka B, Zak I, Krauze J. Synergistic effects of the polymorphisms in the PAI-1 and IL-6 genes with smoking in determining their associated risk with coronary artery disease. Clin Biochem. 2008;41(7–8):467–73. https://doi.org/10.1016/j.clinbiochem.2008.01.028.
Sarecka-Hujar B, Zak I, Krauze J. Carrier-state of two or three polymorphic variants of MTHFR, IL-6 and ICAM1 genes increases the risk of coronary artery disease. Kardiol Pol. 2008;66(12):1269–77.
Aker S, Bantis C, Reis P, Kuhr N, Schwandt C, Grabensee B, et al. Influence of interleukin-6 G-174C gene polymorphism on coronary artery disease, cardiovascular complications and mortality in dialysis patients. Nephrol Dial Transplant. 2009;24(9):2847–51. https://doi.org/10.1093/ndt/gfp141.
Berg KK, Madsen HO, Garred P, Wiseth R, Gunnes S, Videm V. The additive contribution from inflammatory genetic markers on the severity of cardiovascular disease. Scand J Immunol. 2009;69(1):36–42. https://doi.org/10.1111/j.1365-3083.2008.02187.x.
Rios DL, Cerqueira CC, Bonfim-Silva R, Araujo LJ, Pereira JF, Gadelha SR, et al. Interleukin-1 beta and interleukin-6 gene polymorphism associations with angiographically assessed coronary artery disease in Brazilians. Cytokine. 2010;50(3):292–6. https://doi.org/10.1016/j.cyto.2010.02.012.
Bennermo M, Nordin M, Lundman P, Boqvist S, Held C, Samnegard A, et al. Genetic and environmental influences on the plasma interleukin-6 concentration in patients with a recent myocardial infarction: a case-control study. J Interferon Cytokine Res. 2011;31(2):259–64. https://doi.org/10.1089/jir.2010.0036.
Lima-Neto LG, Hirata RD, Luchessi AD, Silbiger VN, Stephano MA, Sampaio MF, et al. CD14 and IL6 polymorphisms are associated with a pro-atherogenic profile in young adults with acute myocardial infarction. J Thromb Thrombolysis. 2013;36(3):332–40. https://doi.org/10.1007/s11239-012-0841-4.
Hatzis G, Tousoulis D, Papageorgiou N, Bouras G, Oikonomou E, Miliou A, et al. Combined effects of smoking and interleukin-6 and C-reactive protein genetic variants on endothelial function, inflammation, thrombosis and incidence of coronary artery disease. Int J Cardiol. 2014;176(1):254–7. https://doi.org/10.1016/j.ijcard.2014.06.058.
Tutun U, Aksoyek A, Ulus AT, Misirlioglu M, Cicekcioglu F, Ozisik K, et al. Gene polymorphisms in patients below 35 years of age who underwent coronary artery bypass surgery. Coron Artery Dis. 2006;17(1):35–9.
Sekuri C, Cam FS, Sagcan A, Ercan E, Tengiz I, Alioglu E, et al. No association of interleukin-6 gene polymorphism (-174 G/C) with premature coronary artery disease in a Turkish cohort. Coron Artery Dis. 2007;18(5):333–7. https://doi.org/10.1097/MCA.0b013e32820588ae.
Ghazouani L, Ben Hadj Khalifa S, Abboud N, Ben Hamda K, Ben Khalfallah A, Brahim N, et al. TNF-alpha -308G>A and IL-6 -174G>C polymorphisms in Tunisian patients with coronary artery disease. Clin Biochem. 2010;43(13–14):1085–9. https://doi.org/10.1016/j.clinbiochem.2010.05.005.
Coker A, Arman A, Soylu O, Tezel T, Yildirim A. Lack of association between IL-1 and IL-6 gene polymorphisms and myocardial infarction in Turkish population. Int J Immunogenet. 2011;38(3):201–8. https://doi.org/10.1111/j.1744-313X.2010.00988.x.
Jabir NR, Firoz CK, Kamal MA, Damanhouri GA, Alama MN, Alzahrani AS, et al. Assessment of genetic diversity in IL-6 and RANTES promoters and their level in Saudi coronary artery disease patients. J Clin Lab Anal. 2016. https://doi.org/10.1002/jcla.22092.
Kuo LT, Yang NI, Cherng WJ, Verma S, Hung MJ, Wang SY, et al. Serum interleukin-6 levels, not genotype, correlate with coronary plaque complexity. Int Heart J. 2008;49(4):391–402.
Fan WH, Liu DL, Xiao LM, Xie CJ, Sun SY, Zhang JC. Coronary heart disease and chronic periodontitis: is polymorphism of interleukin-6 gene the common risk factor in a Chinese population? Oral Dis. 2011;17(3):270–6. https://doi.org/10.1111/j.1601-0825.2010.01736.x.
Li L, Li E, Zhang LH, Jian LG, Liu HP, Wang T. IL-6–174G/C and IL-6–572C/G polymorphisms are associated with increased risk of coronary artery disease. Genet Mol Res. 2015;14(3):8451–7. https://doi.org/10.4238/2015.July.28.12.
Wang K, Dong PS, Zhang HF, Li ZJ, Yang XM, Liu H. Role of interleukin-6 gene polymorphisms in the risk of coronary artery disease. Genet Mol Res. 2015;14(2):3177–83. https://doi.org/10.4238/2015.April.10.29.
Yang HT, Wang SL, Yan LJ, Qian P, Duan HY. Association of interleukin gene polymorphisms with the risk of coronary artery disease. Genet Mol Res. 2015;14(4):12489–96. https://doi.org/10.4238/2015.October.16.16.
Hongmei Y, Yongping J, Jiyuan L. Interleukin-6 polymorphisms and risk of coronary artery diseases in a Chinese population: a case-control study. Pakistan J Med Sci. 2016;32(4):880–5. https://doi.org/10.12669/pjms.324.9908.
Banerjee I, Pandey U, Hasan OM, Parihar R, Tripathi V, Ganesh S. Association between inflammatory gene polymorphisms and coronary artery disease in an Indian population. J Thrombos Thrombolysis. 2009;27(1):88–94. https://doi.org/10.1007/s11239-007-0184-8.
Babu BM, Reddy BP, Priya VH, Munshi A, Rani HS, Latha GS, et al. Cytokine gene polymorphisms in the susceptibility to acute coronary syndrome. Genet Test Mol Biomark. 2012;16(5):359–65. https://doi.org/10.1089/gtmb.2011.0182.
Bhanushali AA, Das BR. Promoter variants in interleukin-6 and tumor necrosis factor alpha and risk of coronary artery disease in a population from Western India. Indian J Human Genet. 2013;19(4):430–6. https://doi.org/10.4103/0971-6866.124371.
Mishra A, Srivastava A, Mittal T, Garg N, Mittal B. Role of inflammatory gene polymorphisms in left ventricular dysfunction (LVD) susceptibility in coronary artery disease (CAD) patients. Cytokine. 2013;61(3):856–61. https://doi.org/10.1016/j.cyto.2012.12.020.
Phulukdaree A, Khan S, Ramkaran P, Govender R, Moodley D, Chuturgoon AA. The interleukin-6 -147 g/c polymorphism is associated with increased risk of coronary artery disease in young South African Indian men. Metab Syndrome Related Disord. 2013;11(3):205–9. https://doi.org/10.1089/met.2012.0130.
Satti HS, Hussain S, Javed Q. Association of interleukin-6 gene promoter polymorphism with coronary artery disease in Pakistani families. Sci World J. 2013;2013:538365. https://doi.org/10.1155/2013/538365.
Biswas S, Ghoshal PK, Mandal N. Synergistic effect of anti and pro-inflammatory cytokine genes and their promoter polymorphism with ST-elevation of myocardial infarction. Gene. 2014;544(2):145–51. https://doi.org/10.1016/j.gene.2014.04.065.
Galimudi RK, Spurthi MK, Padala C, Kumar KG, Mudigonda S, Reddy SG, et al. Interleukin 6(-174G/C) variant and its circulating levels in coronary artery disease patients and their first degree relatives. Inflammation. 2014;37(2):314–21. https://doi.org/10.1007/s10753-013-9742-8.
Ansari WM, Humphries SE, Naveed AK, Khan OJ, Khan DA. Influence of cytokine gene polymorphisms on proinflammatory/anti-inflammatory cytokine imbalance in premature coronary artery disease. Postgraduate Med J. 2016. https://doi.org/10.1136/postgradmedj-2016-134167.
Mitrokhin V, Nikitin A, Brovkina O, Khodyrev D, Zotov A, Vachrushev N, et al. Association between interleukin-6/6R gene polymorphisms and coronary artery disease in Russian population: influence of interleukin-6/6R gene polymorphisms on inflammatory markers. J Inflamm Res. 2017;10:151–60. https://doi.org/10.2147/jir.s141682.
Mastana S, Prakash S, Akam EC, Kirby M, Lindley MR, Sinha N, et al. Genetic association of pro-inflammatory cytokine gene polymorphisms with coronary artery disease (CAD) in a North Indian population. Gene. 2017;628:301–7. https://doi.org/10.1016/j.gene.2017.07.050.
Shabana NA, Ashiq S, Ijaz A, Khalid F, Saadat IU, Khan K, et al. Genetic risk score (GRS) constructed from polymorphisms in the PON1, IL-6, ITGB3, and ALDH2 genes is associated with the risk of coronary artery disease in Pakistani subjects. Lipids Health Dis. 2018;17(1):224. https://doi.org/10.1186/s12944-018-0874-6.
Chen H, Ding S, Liu X, Wu Y, Wu X. Association of interleukin-6 genetic polymorphisms and environment factors interactions with coronary artery disease in a Chinese Han Population. Clin Exp Hypertens. 2018;40(6):514–7. https://doi.org/10.1080/10641963.2017.1403618.
Almeida NCC, Queiroz MAF, Lima SS, Brasil Costa I, Ayin Fossa MA, Vallinoto ACR, et al. Association of Chlamydia trachomatis, C. pneumoniae, and IL-6 and IL-8 gene alterations with heart diseases. Front Immunol. 2019;10:87. https://doi.org/10.3389/fimmu.2019.00087.
Toutouzas K, Klettas D, Anousakis-Vlachochristou N, Melidis K, Azilazian Z, Asimomiti M, et al. The -174 G>C interleukin-6 gene polymorphism is associated with angiographic progression of coronary artery disease in a 4-year period. Hellenic J Cardiol. 2017. https://doi.org/10.1016/j.hjc.2017.02.0020000.
Sepehri Z, Masoumi M, Ebrahimi N, Kiani Z, Nasiri AA, Kohan F, et al. Atorvastatin, losartan and captopril lead to upregulation of TGF-beta, and downregulation of IL-6 in coronary artery disease and hypertension. PLoS ONE. 2016;11(12):e0168312. https://doi.org/10.1371/journal.pone.0168312.
Seino Y, Ikeda U, Ikeda M, Yamamoto K, Misawa Y, Hasegawa T, et al. Interleukin 6 gene transcripts are expressed in human atherosclerotic lesions. Cytokine. 1994;6(1):87–91.
Kishimoto T, Hibi M, Murakami M, Narazaki M, Saito M, Taga T. The molecular biology of interleukin 6 and its receptor. Ciba Foundation Symp. 1992;167:5–16 (discussion -23).
Funding
Open Access funding provided by the IReL Consortium. None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
MJ reports personal fees from Biotronik, personal fees from Orbus Neich, grants and personal fees from Boston Scientific, grants and personal fees from Edwards, personal fees from Recor, personal fees from Astra Zeneca, grants from Amgen, personal fees from Abbott, personal fees from Shockwave, grants from Infraredx, outside the submitted work. RAB reports research funding to the institution from Abbott Vascular, Biosensors, Biotronik and Boston Scientific. All other authors declare no potential conflicts of interest.
Additional information
Handling Editor: John Di Battista.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11_2021_1505_MOESM1_ESM.pptx
Supplementary Figure 1. Forest plot depicting associations of IL6 -174 G/C polymorphism with CAD employing a dominant genetic model (CC+GC vs. GG). Effect sizes for “Pooled” as well as for European and Asian Indian ancestral subgroups displaying a high level of heterogeneity were estimated using random effects for analysis. Effect sizes for Middle Eastern, Asian, African and Mixed ancestral subgroups were estimated using fixed effects. (PPTX 127 KB)
11_2021_1505_MOESM2_ESM.pptx
Supplementary Figure 2. Forest plot depicting associations of IL6 -174 G/C polymorphism with CAD employing an allelic genetic model (Allele C vs. Allele G). Effect sizes for “Pooled” as well as for European and Asian Indian ancestral subgroups displaying high level heterogeneity were estimated using random effects for analysis. Effect sizes for Middle Eastern, Asian, African and Mixed ancestral subgroups were estimated using fixed effects. (PPTX 127 KB)
11_2021_1505_MOESM3_ESM.pptx
Supplementary Figure 3. Forest plots depicting differences in circulating IL-6 levels amongst ‘C’ allele carriers and GG homozygotes. Comparison of IL-6 levels in CC+GC vs. GG genotypes of IL6 -174 G/C polymorphism separately amongst CAD cases and CAD free controls. Standard mean difference for “Pooled” as well as CAD case subgroup were estimated using random effects owing to high levels of inherent heterogeneity. Standard mean difference for CAD free control subgroup which displayed low levels of inherent heterogeneity were estimated using fixed effects. (PPTX 65 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rai, H., Colleran, R., Cassese, S. et al. Association of interleukin 6 -174 G/C polymorphism with coronary artery disease and circulating IL-6 levels: a systematic review and meta-analysis. Inflamm. Res. 70, 1075–1087 (2021). https://doi.org/10.1007/s00011-021-01505-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00011-021-01505-7