Abstract
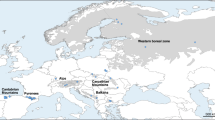
Previous study using protein electrophoresis shows no polymorphism in 44 nuclear loci of Sichuan golden monkey (Rhinopithecus roxellana), which limits our understandings of its population genetic patterns in the nuclear genome. In order to obtain sufficient information, we scanned 14 microsatellite loci in a sample of 32 individuals from its three major habitats (Minshan, Qinling and Shennongjia). A considerable amount of polymorphisms were detected. The average heterozygosities in the local populations were all above 0.5. The differentiations among local populations were significant. There was evidence of geneflow among subpopulations, but geneflow between Qinling and Shennongjia local populations was the weakest. Minshan and Qinling populations might have gone through recent bottlenecks. The estimation of the ratio of the effective population sizes among local populations was close to that from census sizes. Comparisons to available mitochondria data suggested thatR. roxellana’s social structures played an important role in shaping its population genetic patterns. Our study showed that the polymorphism level ofR. roxellana was no higher than other endangered species; therefore, measures should be taken to preserve genetic diversity of this species.
Similar content being viewed by others
References
Quan, G. Q., Xie, J. H., Study of Golden Monkey, Shanghai: Shanghai Science Technology and Education Publishing House, 2002,
Groves, C. P., Primate Taxonomy, in Comparative Evolutionary Biology (ed. D’Araujo, E.), Washington: Smithsonian Institute Press, 2001,
Struhsaker, T. T., Leland, L., Colobines: infanticide by adult males, in Primate Societies (eds. Smuts, B. B., Cheney, D. L., Seyfarth, R. M.), Chicago: University of Chicago Press, 1987, 83–97.
Schaller, G. B., China’s golden treasure, Int.Wildl., 1985, 15:29–31.
Wang, Y. X., Jiang, X. L., Li, D. W., Classification of existing subspecies on golden snub-nosed monkey,Rhinopithecus roxellana (Colobinae, Primates), in Handbook and Absrtracts. XVth Congress of the International Primatological Society, Kuta, 1994, 277.
Ma, S., Wang, Y., The recent distribution, status and conservation of primates in China, Acta Theriologica Sinica, 1988, 8(4): 250–260.
Ren, R. M., Su, Y. J., Yan, K. H. et al., Preliminary survey of the social organization ofRhinopithercus roxellana in shennongjia national natural reserve, Hubei, in The Natural History of the Doucs and Snub-nosed Monkeys (ed. Oxnard C. E., Jablonski N. G.), Singapore: World Scientific Publishing Co. Pte Ltd., 1998, 269–277
Li, B., Pan, R., Oxnard, C. E., Extinction of snub-nosed monkeys in china during the past 400 years, Int. J. Primatol., 2002, 23(6): 1227–1243.[[DOI]
Jablonski, N. G., Dental agenesis as evidence of possible genetic isolation in the colobine monkeyRhinopithecus roxellana, Primates, 1992, 33(3): 371–376.
Li, M., Liang, B., Feng, Z. J. et al., Molecular phylogenetic relationships among Sichuan snub-nosed monkeys (Rhinopithecus roxellanae) Inferred from mitochondrial cytochrome-b gene sequences, Primates, 2001, 42(2): 153–160.
Li, H., Meng, S. J., Men, Z. M. et al., Genetic diversity and population history of golden monkeys (Rhinopithecus roxellana), Genetics, 2003, 164(1): 269–275.
Sambrook, X., Russell, D., Molecular Cloning, New York: Cold Spring Harbor Laboratory Press, 2001,
Morin, P. A., Kanthaswamy, S., Smith, D. G., Simple sequence repeat (ssr) polymorphisms for colony management and population genetics in rhesus macaques (macaca mulatta), Am. J. Primatol., 1997, 42(3): 199–213.[DOI]
Smith, D. G., Kanthaswamy, S., Viray, J. et al., Additional highly polymorphic microsatellite (ssr) loci for estimating kinship in rhesus macaques (Macaca mulatta), Am. J. Primatol., 2000, 50(1): 1–7.[DOI]
Uno, H., Alsum, P., Zimbric, M. L. et al., Colon cancer in aged captive rhesus monkeys (Macaca mulatta), Am. J. Primatol., 1998, 44(1): 19–27.[DOI]
Pritchard, J. K., Feldman, M. W., Statistics for microsatellite variation based on coalescence, Theor. Popul. Biol., 1996, 50(3): 325–344.[DOI]
Cornuet, J. M., Lüikart, G., Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data, Genetics, 1996, 144(4): 2001–2014.
Kalinowski, S. T., How many alleles per locus should be used to estimate genetic distances? Heredity, 2002, 88(1): 62–65.[DOI]
Dieringer, D., Schlötterer, C., Microsatellite analyser (msa): A platform independent analysis tool for large microsatellite data sets, Molecular Ecology Notes, 2003, 3(1): 167–169.[DOI]
Weir, B., Cockerham, C. C., Estimating f-statistics for the analysis of population structure, Evolution, 1984, 38: 1358–1370.
Guo, S. W., Thompson, E. A., Performing the exact test of Hardy-Weinberg proportion for multiple alleles, Biometrics, 1992, 48(2): 361–372.
Rousset, F., Raymond, M., Testing heterozygote excess and deficiency, Genetics, 1995, 140(4): 1413–1419.
Raymond, M., Rousset, F., GENEPOP (version 1.2): Population genetics software for exact tests and ecumenicism, J. Heredity, 1995, 86:248–249.
Schneider, S., Roessli, D., Excoffier, L., Arlequin: A software for population genetics data analysis, Genetics and Biometry Lab, Dept. of Anthropology, University of Geneva, 2000.
Xu, H.Y., Fu, Y. X., Estimating effective population size or mutation rate with microsatellites, Genetics, 2004, 166: 555 -563.[DOI]
Bowcock, A. M., Ruiz-Linares, A., Tomfohrde, J. et al., High resolution of human evolutionary trees with polymorphic microsatellites. Nature, 1994, 368(6470): 455–457.[DOI]
Cavalli-Sforza, L. L., Edwards, A. W. F., Phylogenetic analysis: Models and estimation procedures, Evolution, 1967, 21: 550–570.
Saitou, N., Nei, M., The neighbor-joining method: A new method for reconstructing phylogenetic trees, Mol. Biol. Evol., 1987, 4(4): 406–425.
Felsenstein, J., PHYLIP—Phylogeny inference package (Version 3.2), Cladistics, 2002, 5: 164–166.
Hartl, D. L., Clark, A. G., Principles of Population Genetics, 3rd ed., Sunerland: Sinauer Associates Inc., 1997,
Moran, P.A., Wandering distributions and the electrophoretic profile, Theor. Popul. Biol., 1975, 8(3): 318–330.
Shriver, M. D., Jin, L., Chakraborty, R. et al., VNTR allele frequency distributions under the stepwise mutation model: A computer simulation approach, Genetics, 1993, 134(3): 983–993.
Ota, T., Kimura, M., A model of mutation appropriate to estimate the number of electrophoretically detectable alleles in a finite population, Genet. Res., 1973, 22(2): 201–204.
Zhang, D. X., Hewitt, G. M., Nuclear DNA analyses in genetic studies of populations: Practice, problems and prospects, Mol. Ecol., 2003, 12(3): 563–584.[DOI]
Bennett, P., Demystified microsatellites, Mol. Pathol., 2000, 53(4): 177–183.[DOI]
Lu, Z., Johnson, W. E., Menotti-Raymond, M. et al., Patterns of genetic diversity in remaining Giant Panda populations, Conserv. Biol., 2001, 15(6): 1596–1607.[DOI]
Linda, H., Christopher, W. W., Eli Knispel, R. et al., Differentiation and levels of genetic variation in northern European lynx (Lynx lynx) populations revealed by microsatellites and mitochondrial DNA analysis, Conserv. Genet., 2002, 3(2): 97–111.[DOI]
Zhang, Y. W., Morin, P. A., Ryder, O. A. et al., A set of human triand tetra-nucleotide microsatellite loci useful for population analyses in gorillas (Gorilla gorilla) and orangutans (Pongo pygmaeus), Conserv. Genet., 2001, 2(4): 391–395.[DOI]
Queney, G., Ferrand, N., Weiss, S. et al., Stationary distributions of microsatellite loci between divergent population groups of the European rabbit (Oryctolagus cuniculus), Mol. Biol. Evol., 2001, 18(12): 2169–2178.
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this work.
About this article
Cite this article
Pan, D., Li, Y., Hu, H. et al. Microsatellite polymorphisms of Sichuan golden monkeys. Chin.Sci.Bull. 50, 2850–2855 (2005). https://doi.org/10.1007/BF02899655
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF02899655