Abstract
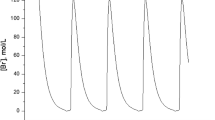
The autonomous oscillations in yeast continuous cultures are investigated analytically and related to the behaviour of the single cell by means of a suitable modified version of Monod’s classical chemostat model. Two main cell phases or states are considered to account for the experimentally observed changes occurring in the cell growth course: the budded phase and the unbudded one. Thus, a sort of two compartment structure is given to the total biomass. The model so far obtained allows one to analyse the local properties of the predicted steady states under various assumptions, both on the yield coefficients and the specific growth rates. Necessary conditions for the local instability are derived and the existence of stable limit cycles is shown by computer simulation. With respect to the qualitative changes in the metabolic parameters, this analysis agrees with the results obtained by simulation of complex structured and segregated models. However, the oscillation period is too long compared with the experimental one and this fact may be mainly due to the strong simplifying assumptions on the dynamic evolution of the transfer rates between the two compartments. The model’s usefulness seems until now restricted to the identification of the relationships between the cell cycle regulation and the oscillation triggering.
Similar content being viewed by others
Abbreviations
- D :
-
dilution rate
- f :
-
mass distribution function
- h :
-
division factor
- k :
-
transfer rate
- m :
-
cell mass
- s :
-
substrate concentration
- Y :
-
yield coefficient
- x :
-
biomass concentration
- ζ:
-
budded biomass fraction
- μ:
-
specific growth rate
- b :
-
budded cells
- c :
-
critical value
- in :
-
input flow
- u :
-
unbudded cells
Literature
Bley, T. 1987. State-structure models of microbial growth.Acta Biotechnol. 7, 173–177.
Carter, B. L. A. and M. N. Jagadish. 1978. The relationship between cell size and cell division in the yeastSaccharomyces cerevisiae.Exp. Cell. Res.,112, 12–24.
Cazzador, L., L. Mariani, E. Martegani and L. Alberghina. 1990. Structured segregated models and analysis of self-oscillating yeast continuous cultures.Bioprocess Engng 5, 175–180.
Hamada, T. and Y. Nakamura. 1982. On the oscillatory transient stage structure of yeast population.J. theor. Biol.,99, 797–805.
Heinzle, E., I. J. Dunn, K. Furukawa and R. D. Tanner. 1982. Modelling of sustained oscillations observed in continuous culture ofSaccharomyces cerevisiae. In:1st IFAC Workshop on Modelling and Control of Biotechnical Processes, IFAC, Helsinki, pp. 57–65.
Hjortso, M. A. and J. E. Bailey. 1983. Transient responses of budding yeast populations.Math. Biosci. 63, 121–148.
Ivanitskaya, J. G., S. B. Petrikevich and A. D. Bazykin. 1989. Oscillations in continuous cultures of microorganisms: criteria of utility of mathematical models.Biotechnol. Bioengng 33, 1162–1166.
Kuenzi, M. T. and A. Fiechter. 1969. Changes in carbohydrate composition and trehalase activity during the budding cycle ofSaccharomyces cerevisiae.Arch. Mikrobiol.,64, 396–407.
Macdonald, N. 1978.Time Lags in Biological Models, Berlin: Springer-Verlag.
Mariani, L., E. Martegani and L. Alberghina. 1986. Yeast population models for monitoring and control of biotechnical processes.IEE Proc.,133, Pt.D. 210–216.
Martegani, E., L. Mariani and L. Alberghina. 1985. Yeast biotechnological processes monitored by analysis of segregated data with structured models. InModelling and Control of Biotechnical Processes, A. Johnson (Ed.), pp. 237–243. Oxford: Pergamon Press.
Parulekar, S. J., G. B. Semoens, M. J. Rolf, J. C. Lievense and H. C. Lim. 1986. Induction and elimination of oscillations in continuous cultures ofSaccharomyces cerevisiae.Biotechnol. Bioengng 28, 700–710.
Porro, D., E. Martegani, B. M. Ranzi and L. Alberghina. 1988. Oscillations in continuous cultures of budding yeast: a segregated parameter analysis.Biotechnol. Bioengng 32, 411–417.
Smith, J. A. and L. Martin 1973. Do cells cycle?Proc. natn. Acad. Sci. U.S.A. 70, 1263–1267.
Strässle, C., B. Sonneleitner and A. Fiechter. 1988. A predictive model for the spontaneous synchronization ofSaccharomyces cerevisiae grown in continuous cultures. I. Concept.J. Biotechnol.,7, 299–318.
von Meyenburg, H. K. 1969. Energetics of the budding cycle ofSaccharomyces cerevisiae during glucose limited aerobic growth.Arch. Mikrobiol. 66, 289–303.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Cazzador, L. Analysis of oscillations in yeast continuous cultures by a new simplified model. Bltn Mathcal Biology 53, 685–700 (1991). https://doi.org/10.1007/BF02461549
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1007/BF02461549