Abstract
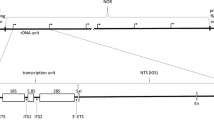
Nucleoli were purified from the slime mold Physarum polycephalum and assayed for enrichment in ribosomal DNA by analytical ultracentrifugation. Greater than 90% of the DNA from nucleoli comprises a satellite band characteristic of Physarum ribosomal DNA (rDNA) in a CsCl equilibrium sedimentation gradient. Over 75% of the nucleolar DNA molecules are 37×106 Daltons, a further characteristic of Physarum rDNA. Nucleoli incubated with micrococcal nuclease yield a distribution of discrete DNA fragments indicative of nucleosome subunits; these digestion products are indistinguishable in size from those of total nuclear chromatin. The nucleosome DNA repeat length varied with increasing digestion from 175 down to 150 base pairs. A palindrome structure for the ribosomal nucleoprotein molecules is demonstrated by electron microscopy of actively transcribing ribosomal RNA genes, which required modification of usual methods for dispersing chromatin. In the center of each palindrome is a 6.0–6.5 μm non-transcribed “spacer” region which exhibits a beaded nucleosome structure. Transcription initiates at points on either side of the central spacer, as evidenced by 4.0–4.2 μm matrices of growing rRNA fibrils extending to each end of the palindrome. The polarity of transcription matrices, together with information about the sites of rRNA coding sequences, imply that 19S rRNA is transcribed prior to 26S rRNA.
Similar content being viewed by others
References
Daniel, J.W., Baldwin, H.H.: Methods of culture for plasmodial myxomycetes. In: Methods in cell physiology (D.M. Prescott, ed.), Vol. 1, pp. 9–41. New York: Academic Press 1964
Davis, R.N., Hyman, R.N.: A study in evolution: the DNA base sequence homology between coli-phages T7 and T3. J. molec. biol. 62, 287–301 (1971)
Davis, R.N., Simon, M., Davidson, N.: Electron microscope heteroduplex methods for mapping regions of base sequence homology in nucleic acids. In: Methods in enzymology (L. Grossman and K. Moldave, eds.), pp. 413–428. New York: Academic Press 1971
Dawid, IIB., Wellauer, P.K.: A reinvestigation of 5′ → 3′ polarity in 40S ribosomal RNA precursor of Xanopus laevis. Cell 8, 443–448. (1976)
Engberg, J., Anderson, P., Leick, V., Collings. J.: Free ribosomal DNA molecules from Tetrahymena pyreformis GL are giant palindromes. J. molec. biol. 104, 455–470 (1976)
Foe, V.W., Wilkinson, C.W., Laird, C.D.: Comparative organization of active transcription units in Oncopeltus fasciatus. Cell 9, 131–146 (1976)
Griffith, J.: Chromatin structure: Deduced from a minichromosome. Science 187, 1202–1203 (1975)
Hackett, P.B., Sauerbier, W.: The transcriptional organization of the ribosomal RNA genes in mouse L cells. J. molec. Biol. 91, 235–256 (1975)
Hall, L., Turnock, G., Cox, B.J.: Ribosomal RNA genes in the amoebal and plasmodial forms of the slime mold Physarum polycephalum. Europ. J. Biochem. 51, 459–465 (1975)
Hamkalo, B., Miller, Jr., O.L.: Electron microscopy of genetic activity. Ann. Rev. Biochem. 42, 379–396 (1973)
Haugli, F.: Mutagenesis, selection and genetic analysis in Physarum polycephalum. Ph. D. Thesis, University of Wisconsin. (1971)
Hewish, D.R., Burgoyne, L.A.: Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem. biophys. Res. Commun. 52, 504–510(1973)
Higashinakagawa, T. Wahn, H., Reeder, R.H.: Isolation of ribosomal gene chromatin. Develop. Biol. 55, 375–386 (1977)
Jacobson, D.N., Holt, C.E.: Isolation of ribosomal RNA precursors from Physarum polycephalum. Arch. Biochem. Biophys. 159, 342–352 (1973)
Jeppeson, P.G.N., Sanders, L., Slocombe, P.: A restriction map of ΦX174 DNA by pulse-chase labelling using E. coli DNA polymerase. Nucleic Acids Res. 3, 1323–1339 (1976)
Johnson, C.M., Littau, V.C., Allfrey, V.G., Bradbury, E.M., Matthews, H.R.: The subunit structure of chromatin from Physarum polycephalum. Nucleic Acids Res. 3, 3313–3329 (1976)
Karrer, D., Gall, J.G.: The macronuclear ribosomal DNA of Tetrahymena pyriformis is a palindrome. J. molec. Biol. 104, 421–453 (1976)
Kornberg, R.: Chromatin structure: A repeating unit of histones and DNA. Science 184, 868–871 (1974)
Laird, C.D., Wilkinson, L.E., Foe, V.W., Chooi, W.Y.: Analysis of chromatin-associated fiber arrays. Chromosoma (Berl.) 58, 169–190 (1976)
Lohr, D., Corden, J., Tatchell, K., Kovacic, R.T., VanHolde, K.E.: Comparative subunit structure of HeLa, Jest, and chicken erythrocyte chromatin. Proc. nat. Acad. Sci. (Wash.) 74, 79–83 (1977)
Mathis, D.J., Gorovsky, M.A.: Subunit structure of rDNA-containing chromatin. Biochemistry 15, 750–755 (1976)
McKnight, S.L., Miller, O.L., Jr.: Ultrastructural patterns of RNA synthesis during early embryogenesis of Drosophila melanogaster. Cell 8, 305–319 (1976)
McKnight, S.L., Miller, Jr. O.L.: Electron microscopic analysis of replicating Drosophila melanogaster chromatin: Evidence for rapid nucleosome regeneration. Cell (in press, 1977)
Melera, P.W., Rusch, H.P.: A characterization of ribonucleic Acid in the myxomycete Physarum polycephalum. Exp. Cell Res. 82, 197–209 (1973)
Miller, Jr., O.L., Beatty, B.: Visualization of nucleolar genes. Science 164, 955–957 (1969)
Mohberg, J., Rusch, H.P.: Isolation and DNA content of nuclei of Physarum polycephalum. Exp. Cell Res. 66, 305–316 (1971)
Molgaard, H.V., Matthews, H.R., Bradbury, E.M.: Organization of genes for ribosomal RNA in Physarum polycephalum. Europ. J. Biochem. 68, 541–549 (1976)
Newlon, C., Sonenshein, G., Holt, C.: Time of synthesis of genes for ribosomal ribonucleic acid in Physarum. Biochemistry 12, 2338–2345 (1973)
Noll, M.: Subunit structure of chromatin. Nature (Lond.) 251, 249–251 (1974)
Olins, D.E., Olins, A.L.: Spheroid chromatin units (v-bodies). Science 183, 330–332 (1974)
Oudet, P., Gross-Bellard, M., Chambon, P.: Electron microscopic and biochemical evidence that chromatin structure is a repeating unit. Cell 4, 281–300 (1975)
Perry, R.: Processing of RNA. Ann. Rev. Biochem. 45, 605–629 (1976)
Reeder, R.H., Higashinakagawa, R., Miller, Jr., O.L.: The 5′ → 3′ polarity of the Xenopus ribosomal RNA precursor molecule. Cell 8, 449–454 (1976)
Reeves, R.: Ribosomal genes of Xenopus laevis: Evidence of nucleosomes in transcriptionally active chromatin. Science 194, 529–532 (1976)
Ritossa, F.M., Spiegelman, S.: Localization of DNA complementary to ribosomal RNA in the nucleoulus organizer region of Drosophila melanogaster. Proc. nat. Acad. Sci. (Wash.) 53, 737–745 (1965)
Shaw, B.R., Herman, T.M., Kovacic, R.T., Beaudreau, G.S., Van Holde, K.E.: Analysis of subunit organization in chicken erythrocyte chromatin. Proc. nat. Acad. Sci. (Wash.) 73, 505–509 (1976)
Solner-Webb, B., Felsenfeld, G.: A comparison of the digestion of nuclei and chromatin by staphylococcal nuclease. Biochemistry 14, 2915–2920 (1975)
Steele, W.J.: Localization of deoxyribonucleic acid complementary to ribosomal ribonucleic acid and preribosomal ribonucleic acid in the nucleolus of rat liver. J. biol. Chem. 243, 3333–3341 (1968)
Sugden, B., DeTroy, B., Roberts, R.J., Sambrook, J.: Agarose slab-gel electrophoresis equipment. Anal. Biochem. 68, 36–46 (1975)
Thomas, J.O., Furber, V.: Yeast chromatin structure. FEBS Letters 66, 274–280 (1976)
Vogt, V., Braun, R.: The structure of ribosomal DNA in Physarum polycephalum. J. molec. Biol. 106, 567–587 (1976)
Wallace, H., Birnstiel, M.L.: Ribosomal cistrons and the nucleolus organizer. Biochim. biophys. acta (Amst.) 114, 296–310 (1966)
Weintraub, H., Groudine, M.: Chromosomal subunits in active genes have an altered conformation. Science 193, 848–856 (1976)
Woodcock, C.L.F., Safer, J.P., Stanchfield, J.E.: Structural Repeating units in chromatin. Exp. Cell Res. 97, 101–110 (1976)
Yao, M.C., Gall, J.G.: A single integrated gene for ribosomal RNA in a eukaryote, Tetrahymena pyriformis. Cell (in press 1977)
Zellweger, A., Ryser, U., Braun, R.: Ribosomal genes of Physarum: Their isolation and replication in the mitotic cycle. J. molec. Biol. 64, 681–691 (1972)
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Grainger, R.M., Ogle, R.C. Chromatin structure of the ribosomal RNA genes in Physarum polycephalum . Chromosoma 65, 115–126 (1978). https://doi.org/10.1007/BF00329464
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1007/BF00329464