Abstract
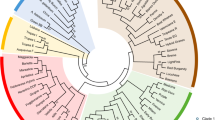
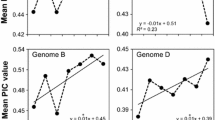
Sequencing the complete genome of Camelina sativa will facilitate studies to improve this oilseed crop. We analysed 20 accessions of Camelina spp. using genotyping-by-sequence technology. After stringent screening, 35,783 single nucleotide polymorphisms (SNPs) were generated, and basic genetic studies were performed to check the diversity of these SNPs. STRUCTURE and phylogenetic analyses revealed five subgroups in these 20 Camelina accessions. Winter-types may have diverged from summer-types. Some genomic regions were negatively selected, and most of these were gene-rich regions. As expected, the most ancient subgroup was less affected by negative selection. Marker–trait associations with plant height, leaf length, and pod size generated 154 SNPs, and 72 adjacent genes were significantly associated with these phenotypes. Further large-scale analysis and gene expression studies with these SNPs and genes are needed to develop potentially valuable resources for improving C. sativa.
Similar content being viewed by others
Literature Cited
Agegnehu M, Honermeier B (1997) Effects of seeding rates and nitrogen fertilization on seed yield, seed quality and yield components of false flax (Camelina sativa Crtz). Bodenkultur 48:15–21
Akeroyd JR (1993) Camelina Crantz. Cambridge University Press, Cambridge
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23:2633–2635
Brooks RE (1985) Chromosome number reports. LXXXVII. Taxon 34:346–351
Evanno G, S. R, J. G (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Francis A, Warwick SI (2009) The biology of Canadian weeds. 142. Camelina alyssum (Mill.) Thell.; C. microcarpa Andrz. ex DC.; C. sativa (L.) Crantz. Can J Plant Sci 89:791–810
Gehringer A, Friedt W, Luhs W, Snowdon RJ (2006) Genetic mapping of agronomic traits in false flax (Camelina sativa subsp. sativa). Genome 49:1555–1563
Ghamkhar K, Croser J, Aryamanesh N, Campbell M, Kon’kova N, Francis C (2010) Camelina (Camelina sativa (L.) Crantz) as an alternative oilseed: molecular and ecogeographic analyses. Genome 53:558–567
Glaubitz JC, Casstevens TM, Lu F, Harriman J, Elshire RJ, Sun Q, Buckler ES (2014) TASSEL-GBS: a high capacity genotyping by sequencing analysis pipeline. PLoS ONE 9:e90346
Hutcheon C, Ditt RF, Beilstein M, Comai L, Schroeder J, Goldstein E, Shewmaker CK, Nguyen T, De Rocher J, et al. (2010) Polyploid genome of Camelina sativa revealed by isolation of fatty acid synthesis genes. BMC Plant Biol 10:233
Kagale S, Koh C, Nixon J, Bollina V, Clarke WE, Tuteja R, Spillane C, Robinson SJ, Links MG, et al. (2014) The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat Commun 5:3706
Keller B, Feuillet C (2000) Colinearity and gene density in grass genomes. Trends Plant Sci 5:246–251
Kim B, Kim N, Kang J, Choi JY, Sim S-C, Min SR, Park Y (2015) Single Nucleotide Polymorphisms linked to the SlMYB12 Gene that Controls Fruit Peel Color in Domesticated Tomatoes (Solanum lycopersicum L.). Korean J Hortic Sci Technol 33:566–574
Larsson SJ, Lipka AE, Buckler ES (2013) Lessons from Dwarf8 on the strengths and weaknesses of structured association mapping. PLoS Genet 9:e1003246
Lee TH, Guo H, Wang X, Kim C, Paterson AH (2014) SNPhylo: a pipeline to construct a phylogenetic tree from huge SNP data. BMC Genomics 15:162
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Liang C, Liu X, Yiu SM, Lim BL (2013) De novo assembly and characterization of Camelina sativa transcriptome by paired-end sequencing. BMC Genomics 14:146
Librado P, Rozas J (2009) DnaSP v5:a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Lin X, Kaul S, Rounsley S, Shea TP, Benito MI, Town CD, Fujii CY, Mason T, Bowman CL, et al. (1999) Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402:761–768
Maassoumi A (1980) Cruciferes de la flore d’Iran: etude caryosystematique, Strasbourg, France.
Mudalkar S, Golla R, Ghatty S, Reddy AR (2014) De novo transcriptome analysis of an imminent biofuel crop, Camelina sativa L. using Illumina GAIIX sequencing platform and identification of SSR markers. Plant Mol Biol 84:159–171
Nguyen HT, Silva JE, Podicheti R, Macrander J, Yang W, Nazarenus TJ, Nam JW, Jaworski JG, Lu C, et al. (2013) Camelina seed transcriptome: a tool for meal and oil improvement and translational research. Plant Biotechnol J 11:759–769
Nordborg M, Borevitz JO, Bergelson J, Berry CC, Chory J, Hagenblad J, Kreitman M, Maloof JN, Noyes T, et al. (2002) The extent of linkage disequilibrium in Arabidopsis thaliana. Nat Genet 30:190–193
Poland JA, Brown PJ, Sorrells ME, Jannink JL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 7:e32253
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Wang S, Dvorkin D, Da Y (2012) SNPEVG: a graphical tool for GWAS graphing with mouse clicks. BMC Bioinformatics 13:319
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this work.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kim, C., Lee, J.H., Chung, Y.S. et al. Characterization of twenty Camelina spp. accessions using single nucleotide polymorphism genotyping. Hortic. Environ. Biotechnol. 58, 187–194 (2017). https://doi.org/10.1007/s13580-017-0264-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13580-017-0264-4