Abstract
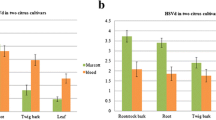
Citrus tristeza virus (CTV) isolates were detected in Poncirus trifoliata, but few research focused on the distribution and population structure of CTV isolates in this resistant species. In this study, 9 P. trifoliata cv. ‘Zaoyangxiaoye’ seedlings were tested positive for CTV in the field. CTV was detected in petal, stamen, pistil and rootlet tissues from CTV-infected plants by real-time RT-PCR, but the incidence of CTV in young flush and old shoot tissues was 68.9% and 4.4%, respectively. The results also showed the highest relative amount of CTV was detected in pistils, followed by rootlets, stamens, petals and young flush. The lowest CTV accumulation was in old shoots, with a viral load two orders of magnitude lower than in pistils. Furthermore, CTV present at a concentration approximately 5 to 10 times higher in petals than in young shoots. These results indicated that the floral parts and rootlets of Zaoyangxiaoye trifoliate orange were more susceptible to CTV accumulation than shoots. Furthermore, T36, T30 and VT genotypes were detected in all of CTV-infected petal, stamen, pistil, rootlet and young flush tissues, but only VT genotype was found in old shoots. Further analysis by specific real-time RT-PCR suggested that the relative amount of VT genotype was most abundant in all of tissue samples, followed by the T30 and T36 genotypes.
Similar content being viewed by others
References
Albiach-Marti MR, Grosser JW, Gowda S, Mawassi M, Satyanarayana T, Garnsey SM, Dawson WO (2004) Citrus tristeza virus replicates and forms infections virions in protoplasts of resistant citrus relatives. Mol Breed 14:117–128
Ananthakrishnan G, Venkataprasanna T, Roy A, Brlansky RH (2010) Characterization of the mixture of genotypes of a Citrus tristeza virus isolate by reverse transcription quantitative real-time PCR. J Virolo Methods 164:75–82
Andika IB, Kondo H, Nishiguchi M, Tamada T (2012) The cysteine-rich proteins of Beet necrotic yellow vein virus and Tobacco rattle virus contribute to efficient suppression of silencing in roots. J Gen Virol 93:1841–1850
Bar-Joseph M, Marcus R, Lee RF (1989) The continuous challenge of Citrus tristeza virus control. Annu Rev Phytopathol 27:291–316
Cook G, van Vuuren SP, Breytenbach JHJ, Burger JT, Maree HJ (2015) Expanded strain-specific RT-PCR assay for differential detection of currently known Citrus tristeza virus strains: a useful screening tool. J Phytopathol 164:847–851
Dawson TE, Mooney PA (2000) Evidence for trifoliate resistance breaking isolates of Citrus tristeza virus in New Zealand. In: da Graça JV, Lee RF, Yokomi RK (eds) Proceedings of the 14th conference of the International Organization of Citrus Virologist. IOCV, Riverside, pp 69–76
Deng Z, Haung S, Xiao S, Gmitter FG (1997) Development and characterization of SCAR markers linked to the Citrus tristeza virus resistance gene from Poncirus trifoliata. Genome 40:697–704
Dimitrov DS (2004) Virus entry: molecular mechanisms and biomedical applications. Nat Rev Microbiol 2:109–122
Fagoaga C, Pensabene-Bellavia G, Moreno P, Navarro L, Flores R, Peña L (2011) Ectopic expression of the p23 silencing suppressor of Citrus tristeza virus differentially modifies viral accumulation and tropism in two transgenic woody hosts. Mol Plant Pathol 12:898–910
Folimonova SY, Folomonov AS, Satyanarayana T, Dawson T (2008) Citrus tristeza virus: survival at the edge of the movement continuum. J Virol 82:6546–6556
Gómez-Muñoz N, Velázǫuez K, Vives MC, Ruiz-Ruiz S, Pina J, Flores R, Moreno P, Guerri J (2016) The resistance of sour orange to Citrus tristeza virus is mediated by both the salicylic acid and RNA silencing defence pathways. Mol Plant Pathol. doi:10.1111/mpp.12488
Harper SJ (2013) Citrus tristeza virus: evolution of complex and varied genotypic groups. Front Microbiol. doi:10.3389/fmicb.2013.00093
Harper SJ, Dawson TE, Pearson MN (2009) Complete genome sequences of two distinct and diverse Citrus tristeza virus isolates from New Zealand. Arch Virol 154:1505–1510
Harper SJ, Dawson TE, Pearson MN (2010) Isolates of Citrus tristeza virus that overcome Poncirus trifoliata resistance comprise a novel strain. Arch Virol 155:471–480
Harper SJ, Cowell SJ, Robertson CJ, Dawson WO (2014) Differential tropism in roots and shoots infected by Citrus tristeza virus. Virology 460-461:91–99
Harper SJ, Cowell SJ, Dawson WO (2015) With a little help from my friends: complementation as a survival strategy for viruses in a long-lived host system. Virology 478:123–128
Hilf ME, Mavrodieva VA, Garnsey SM (2005) Genetic marker analysis of a global collection of isolates of Citrus tristeza virus: characterization and distribution of CTV genotypes and association with symptoms. Phytopathology 95:909–917
Kleynhans J, Pietersen G (2016) Comparison of multiple viral population characterization methods on a candidate cross-protection Citrus tristeza virus (CTV) source. J Virolo Methods 237:92–100
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408
Manabayeva SA, Shamekova M, Park JW, Ding XS, Nelson RS, Hsieh YC, Omarov RT, Scholthof HB (2013) Differential requirements for Tombusvirus coat protein and p19 in plants following leaf versus root inoculation. Virology 439:89–96
Martín-Hernández AM, Baulcombe DC (2008) Tobacco rattle virus 16-kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. J Virol 82:4064–4071
Moreno P, Ambros S, Albiach-Marti MR, Guerri J, Pena L (2008) Citrus tristeza virus: a pathogen that changed the course of the citrus industry. Mol Plant Pathol 9:251–268
Roy A, Ananthakrishnan G, Hartung JS, Brlansky RH (2010) Development and application of a multiplex reverse-transcription polymerase chain reaction assay for screening a global collection of Citrus tristeza virus isolates. Phytopathology 100:1077–1088
Ruiz-Ruiz S, Moreno P, Guerri J, Ambrós S (2007) A real-time RT-PCR assay for detection and absolute quantitation of Citrus tristeza virus in different plant tissues. J Virol Methods 145:96–105
Ruiz-Ruiz S, Soler N, Sánchez-Navarro J, Fagoaga C, López C, Navarro L, Moreno P, Peña L, Flores R (2013) Citrus tristeza virus p23: determinants for nucleolar localization and their influence on suppression of RNA silencing and pathogenesis. Mol Plant Microbe In 26:306–318
Yokomi RK, Selvaraj V, Maheshwari Y, Saponari M, Giampetruzzi A, Chiumenti M, Hajeri S (2017) Identification and characterization of Citrus tristeza virus isolates breaking resistance in trifoliate orange in California. Phytopathology. doi:10.1094/PHYTO-01-17-0007-R
Zhou CY, Zhao XY, Jiang YH, Yang FY, Yang KZ, Li TS, Dai SG (1996) Characterization of Citrus tristeza virus isolates infecting pummelo and sweet orange in Sichuan province, China. In: da Graça JV, Moreno P, Yokomi RK (eds) In: proceedings of the 13th conference of the International Organization of Citrus Virologist. IOCV, Riverside, pp 78–82
Zhou Y, Zhou CY, Li ZA, Wang XF, Liu KH (2008) Mild strains cross protection against stem-pitting tristeza of sweet orange. Sci Agric Sin 41:4085–4091
Acknowledgments
This work was partially supported by Chongqing Research Program of Basic Research, and Frontier Technology (cstc2015jcyjBX0043), Fundamental Research Funds for the Central Universities (XDJK2015A009, XDJK2013C103) and Startup Foundation for Doctors of Southwest University (swu113102).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, Y., Liu, Y., Liu, K. et al. Distribution and population structure of Citrus tristeza virus in Poncirus trifoliata . Australasian Plant Pathol. 46, 351–355 (2017). https://doi.org/10.1007/s13313-017-0498-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-017-0498-8