Abstract
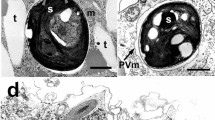
The process of endosymbiosis is one of the promoters of speciation. The green hydra (Hydra viridissima Pallas, 1766) is a typical example of endosymbiosis. Its gastrodermal myoepithelial cells harbour the individuals of unicellular photoautotrophic algae. In this study we have developed healthy laboratory cultures of endosymbiotic green algae isolated from green hydra strains. The Hydra strains were collected from four different geographical localities, two of which were in Croatia (strains BV, T), one in Israel (strain M9) and one in Germany (strain HV). For the first time, endosymbiotic algae isolated from green hydra strains have been visualized and described by scanning and transmission electron microscopy. Isolated endosymbiotic algae were characterized by cytological morphometric parameters, enzyme activity and isoenzyme pattern analysis (catalase, peroxidase and esterase). In addition, cells of endosymbiotic algae were characterized by morphometric measurements of diameter, perimeter and area, and chloroplast area. Endosymbiotic algae HV (collected in Germany) and M9 (collected in Israel) were significantly different when compared with endosymbiotic algae T and BV collected in Croatia. The endosymbionts were more different with referent alga Parachlorella kessleri than with referent alga Chlorella vulgaris. The results obtained by isoenzyme pattern analysis also suggested that there was a difference between Croatian and European algal endosymbionts, i.e. the results indicate biological diversity among algal symbionts isolated from the green hydras from different geographical localities. Ultimately, five isoenzymes of catalase and five isoenzymes of peroxidase were resolved by PAGE electrophoresis. Catalase isoenzyme K1 appeared only in endosymbiotic alga M9 collected in Israel and catalase isoenzyme K3 only in endosymbiotic alga T collected in Croatia. Peroxidase isoenzyme P1 was observed only in endosymbiont HV while peroxidase isoenzyme P5 was observed in both endosymbionts HV and M9. Peroxidase isoenzymes P3 and P4 were visible in endosymbiotic algae BV and T collected in Croatia.










Similar content being viewed by others
References
Aebi H (1984) Catalase in vitro. Methods Enzymol 105:121–126
Balen B, Krsnik-Rasol M, Simeon-Rudolf V (2003) Isoenzymes of peroxidase and esterase related to morphogenesis in Mammillaria gracilis Pfeiff. tissue culture. J Plant Physiol 160:1401–1406
Beijerinck MW (1890) Culturversuche mit Zoochlorellen, Lichenene-gonidien und anderen niederen Algen I-III. Bot Ztg 48:726–740
Bradford MM (1976) A rapid and sensitive assay for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Burlina A, Galzigna L (1972) A new and simple procedure for serum arylesterase. Clin Chim Acta 39:255–257
Burlina A, Michielin E, Galzigna L (1999) Characteristics and behaviour of arylesterase in human serum and liver. Eur J Clin Investig 7:17–20
Chance B, Maehly AC (1955) Assay of catalases and peroxidases. In: Colowick SP, Kaplan NO (eds) Methods in enzymology. Academic Press, New York, pp 764–775
Chelikani P, Fita I, Loewen PC (2004) Diversity of structures and properties among catalases. Cell Mol Life Sci 61:192–208
Collins AG, Schuchert P, Marques AC, Jankowski T, Medina M, Schierwate B (2006) Medusozoan phylogeny and character evolution clarified by new large and small subunit rDNA data and an assessment of the utility of phylogenetic mixture models. Syst Biol 55:97–115
Douglas AE (1994) Symbiotic interactions. Oxford University Press, New York
Dunahay TG, Jarvis EE, Zeiler KG, Roessler PG, Brown LM (1992) Genetic engineering of microalgae for fuel production. Appl Biochem Biotechnol 34(35):331–339
Falkowski PG, Raven JA (1997) Aquatic photosynthesis. Blackwell Scientific, Oxford
Felsenstein J (1975) The genetic basis of evolutionary change (1975). Evolution 29:587–590
Galliot B, Schmid V (2002) Cnidarians as a model system for understanding evolution and regeneration. Int J Dev Biol 46:39–48
Gaspar T, Penel C, Hagege D, Greppin H (1991) Peroxidases in plant growth, differentiation and developmental processes. In: Lobarzewski J, Greppin H, Penel C, Th G (eds) Biochemical, molecular and physiological aspects of plant peroxidases. University M. Curie-Skłodowska. University of Geneva, Lublin, Geneva, pp 249–280
Graham LE, Graham JM, Wilcox LW (2009) Algae, 2nd edn. Pearson Education, San Francisco
Habetha M, Anton-Erksleben F, Neumann K, Bosch TCG (2003) The Hydra viridis/Chlorella symbiosis. Growth and sexual differentiation in polyps without symbionts. Zoology 106:1–8
Hoshina R, Imamura N (2008) Multiple origins of the symbioses in Paramecium bursaria. Protist 159:53–63
Huss VAR, Holweg C, Seidel B, Reich V, Rahat M, Kessler E (1993/1994) There is an ecological basis for host/symbiont specificity in Chlorella/Hydra symbioses. Endocytobiosis. Cell Res 10:35–46
Kawaida H, Ohba K, Koutake Y, Shimizu H, Tachida H, Kobayakawa Y (2013) Symbiosis between hydra and chlorella: molecular phylogenetic analysis and experimental study provide insight into its origin and evolution. Mol Phylogenet Evol 66:906–914
Kessler E, Huss VAR (1992) Comparative physiology and biochemistry and taxonomic assignment of the Chlorella (Chlorophyceae) strains of the culture collection of the University of Texas at Austin. J Phycol 28:550–553
Kovačević G, Franjević D, Jelenčić B, Kalafatić M (2010a) Isolation and cultivation of endosymbiotic algae from green hydra and phylogenetic analysis of 18S rDNA sequences. Folia Biol (Kraków) 58:135–143
Kovačević G, Radić S, Jelenčić B, Kalafatić M, Posilović H, Pevalek-Kozlina B (2010b) Morphological features and isoenzyme characterization of endosymbiotic algae from green hydra. Plant Syst Evol 284:33–39
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Laloue H, Weber-Lotfi F, Lucau-Danila A, Guillemaut P (1997) Identification of ascorbate and guaiacol peroxidases in needle chloroplasts of spruce trees. Plant Physiol Bioch 35:341–346
Lebeda A, Luhová L, Sedlářová M, Jančová D (2001) The role of enzymes in plant-fungal pathogens interactions. Z Pflanzenk Pflanzen 108:89–111
Leliaert F, Verbruggen H, Zechman FW (2011) Into the deep: new discoveries at the base of the green plant phylogeny. BioEssays 33:683–692
Lewis LA, Muller-Parker G (2004) Phylogenetic placement of "zoochlorellae" (Chlorophyta), algal symbiont of the temperate sea anemone Anthopleura elegantissima. Biol Bull 207:87–92
Mittler R, Zilinskas B (1993) Detection of ascorbate peroxidase activity in native gels by inhibition of the ascorbate-dependent reduction of nitroblue tetrazolium. Anal Biochem 212:540–546
Miyazawa Y, Murayama T, Ooya N, Wang LF, Tung YC, Yamaguchi N (1988) Immunomodulation by a unicellular green algae (Chlorella pyrenoidosa) in tumor-bearing mice. J Ethnopharmacol 24:135–146
Pardy RL (1983) Preparing aposymbiotic hydra. In: Lenhoff HM (ed) Hydra: research methods. Plenum Press, New York, pp 394–395
Pröschold T, Darienko T, Silva PC, Reisser W, Krienitz L (2011) The systematics of Zoochlorella revisited employing an integrative approach. Environ Microbiol 13:350–364
Rahat M (1991) An ecological approach to hydra-cell colonization by algae-algae/hydra symbioses. Oikos 62:381–388
Rahat M, Reich V (1986) Algal endosymbiosis in brown hydra: host/symbiont specificity. J Cell Sci 86:273–286
Rajević N, Kovačević G, Kalafatić M, Gould SB, Martin WF, Franjević D (2015) Algal endosymbionts in European Hydra strains reflect multiple origins of the zoochlorella symbiosis. Mol Phylogenet Evol 93:55–62
Reynolds ES (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol 17:208–212
Samec P, Posvec Z, Stejskal J, Nasinec V, Griga M (1998) Cultivar identification and relationship in Pisum sativum L. based on RAPD and isozymes. Biol Plantarum 41:39–48
Sharma P, Jha AB, Dubey RS, Pessarakli M (2012) Reactive oxygen species, oxidative damage, and antioxidative defense mechanism in plants under stressful conditions. J Botany 2012, Article ID e217037, 26 pages. https://doi.org/10.1155/2012/217037
Tanaka K, Koga T, Konishi F, Nakamura M, Mitsuyama M, Himeno K, Nomoto K (1986) Augmentation of host defense by a unicellular green alga, Chlorella vulgaris, to Escherichia coli infection. Infect Immun 53:267–271
Technau U, Steele RE (2011) Evolutionary crossroads in developmental biology: Cnidaria. Development 138:1447–1458
Woodbury W, Spencer AK, Stahmann MA (1971) An improved procedure using ferrycyanide for detecting catalase isozymes. Anal Biochem 44:301–305
Acknowledgements
Very special thanks to Ms. Ivana Rajević for technical support with photographs and to Prof. Ivana Bočina for proofreading the manuscript. This work was supported by Adris project and Ministry of Science, Education and Sport of the Republic of Croatia project number 119-1193080-1214 “Molecular phylogeny, evolution and symbiosis of freshwater invertebrates”.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
S1
(a-b) Laboratory cultivation of green hydra strains in Division of Zoology, Department of Biology, Faculty of Science, University of Zagreb (PNG 3371 kb)
S2
Endosymbiotic algae isolated from different green Hydra hosts in growing cultures. (a) Endosymbiotic alga isolated from green hydra strain T (b) Endosymbiotic alga isolated from green hydra strain BV (c) Endosymbiotic alga isolated from green hydra strain M9 (d) Endosymbiotic alga isolated from green hydra strain HV. Scale bar 1 cm (PNG 7866 kb)
Rights and permissions
About this article
Cite this article
Kević, N., Brkanac, S.R., Vincek, N. et al. Endosymbiotic green algae in European Hydra strains show quantitative difference on morphological and isoenzyme level. Symbiosis 77, 161–175 (2019). https://doi.org/10.1007/s13199-018-0579-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-018-0579-7