Abstract
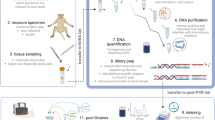
Carnivores play an important role on the ecosystem equilibrium. However, due to habitat loss and fragmentation, their populations have been declining around the world. Together, the elusive behavior and often low population density challenge the data collection. In this way, non-invasive sampling has been considered as an important alternative for information gathering without need of animal capture. Molecular analysis has been essential for precise species identification through non-invasive samples, as scats. However, as DNA from these samples is usually found degraded, the use of more than one marker for a safer identification is critical. In this work, we tested a set of new mini barcoding primers for molecular identification of non-invasive samples for Neotropical felid and canid species. Primers for three mitochondrial DNA regions—rRNA12Ss1, rRNA12Ss2, rRNA16S—were designed for canid and felid species and amplified using fecal samples, while control region and CytB primers were designed only for canids, totaling five mini barcoding primer pairs successfully tested in fecal samples. This study resulted in a very efficient primer set, which represents an important tool for a precise identification of South American carnivores based on non-invasive samples.
Similar content being viewed by others
References
Ausband DE et al (2014) Monitoring gray wolf populations using multiple survey methods. J Wildl Manag 78:335–346. https://doi.org/10.1002/jwmg.654
Ball MC et al (2007) Characterization of target nuclear DNA from faeces reduces technical issues associated with the assumptions of low-quality and quantity template. Conserv Genet 8:577–586. https://doi.org/10.1007/s10592-006-9193-y
Balme GA, Hunter LTB, Slotow R (2009) Evaluating methods for counting cryptic carnivores. J Wildl Manag 73:433–441
Barea-Azcón JM, Virgós E, Ballesteros-Duperon E, Moleón M, Chirosa M (2007) Surveying carnivores at large spatial scales: a comparison of four broad-applied methods. Biodivers Conserv 16:1213–1230. https://doi.org/10.1007/s10531-006-9114-x
Bellemain E, Taberlet P (2004) Improved noninvasive genotyping method: application to brown bear (Ursus arctos) faeces. Mol Ecol Notes 4:519–522. https://doi.org/10.1111/j.1471-8286.2004.00711.x
Benson DA, Clark K, Karsch-Mizrachi I et al (2014) GenBank Nucleic Acids Res 42:D32–D37
Caragiulo A et al (2014) Mitochondrial DNA sequence variation and phylogeography of Neotropic pumas (Puma concolor). Mitochondrial DNA 25:304–312
Chaves PB, GraefF VG, Lion MB, Oliveira LR, Eizirik E (2012) DNA barcoding meets molecular scatology: short mtDNA sequences for standardized species assignment of carnivore noninvasive samples. Mol Ecol Res 12:18–35
Costa LP, Leite YLR, Mendes SL, Ditchfield AD (2005) Mammal conservation in Brazil Conserv Biol 19:672–679
Crooks KR (2002) Relative sensitivities of mammalian carnivores to habitat fragmentation. Conserv Biol 16:488–502
Dalén L, Götherström A, Angerbjörn A (2004) Identifying species from pieces of faeces. Conserv Genet 5:109–111
De Barba M, Adams JR, Goldberg CS, Stansbury CR, Arias D, Cisneros R, Waits LP (2014) Molecular species identification for multiple carnivores. Conserv Genet Res 6:821–824. https://doi.org/10.1007/s12686-014-0257-x
Dos Reis NR, Peracchi AL, Pedro WA, Lima IP (2011) Mamíferos do Brasil; Mammals of Brazil. Segunda edn. Universidade Estadual de Londrina, Londrina
Estes JA et al (2011) Trophic downgrading of planet Earth Science 333:301–306. https://doi.org/10.1126/science.1205106
Farrell LE, Roman J, Sunquist ME (2000) Dietary separation of sympatric carnivores identified by molecular analysis of scats. Mol Ecol 9:1583–1590
Fernandes CA, Ginja C, Pereira I, Tenreiro R, Bruford MW, Santos-Reis M (2008) Species-specific mitochondrial DNA markers for identification of non-invasive samples from sympatric carnivores in the Iberian Peninsula Conservn Genet 9:681–690. https://doi.org/10.1007/s10592-007-9364-5
IUCN (2016) The IUCN Red List of Threatened Species. International Union for Conservation of Nature and Natural Resources. http://www.iucnredlist.org/
Johnson WE et al (2006) The late Miocene radiation of modern Felidae: a genetic assessment. Science 311:5757, 73–77
Kearse M et al (2012) Geneious Basic: an integrated and extendable desktop software platform for the organization analysis of sequence data Bioinformatics 28:1647–1649
Kitano T, Umetsu K, Tian W, Osawa M (2007) Two universal primer sets for species identification among vertebrates. Int J Legal Med 121:423–427
Kocher TD et al (1989) Dynamics of mitochondrial DNA evolution in animals: amplification and sequencing with conserved primers. Proc Natl Acad Sci USA 86:6196–6200
Lindblad-Toh K et al (2005) Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438:803–819
Owczarzy R et al (2008) IDT SciTools: a suite for analysis and design of nucleic acid oligomers. Nucleic Acids Res 36:W163-W169
Palomares F, Godoy JA, Píriz A, O’Brien SJ (2002) Faecal genetic analysis to determine the presence and distribution of elusive carnivores: design and feasibility for the Iberian. lynx Mol Ecol 11:2171–2182
Ray J, Redford KH, Steneck R, Berger J (2013) Large carnivores and the conservation of biodiversity. Island Press, Washington
Ripple WJ, Beschta RL (2012) Large predators limit herbivore densities in northern forest ecosystems European. Journal of Wildlife Research 58:733–742
Ripple WJ et al (2014) Status and ecological effects of the world’s largest Carnivores Science 343:1241484. https://doi.org/10.1126/science.1241484
Rodgers TW, Janečka JE (2013) Applications and techniques for non-invasive faecal genetics research in felid conservation European. J Wildl Res 59:1–16. https://doi.org/10.1007/s10344-012-0675-6
Rozen S, Skaletsky HJ (2000) Primer3 on the WWW for general users and for biologist programmers. In Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press, Totowa, NJ, pp 365–386
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2. Cold Spring Harbor Laboratory Press, New York
Saranholi BH, Bergel MM, Ruffino PHP, Rodríguez-C KG, Ramazzotto LA, de Freitas PD, Galetti PM Jr (2016) Roadkill hotspots in a protected area of Cerrado in Brazil: planning actions to conservation Rev MVZ Córdoba 21 5441–5448
Schmidt JH, Rattenbury KL, Robison HL, Gorn TS, Shults BS (2017) Using non-invasive mark-resight and sign occupancy surveys to monitor low-density brown bear populations across large landscapes. Biol Cons 207:47–54
Shehzad W et al (2012) Carnivore diet analysis based on next-generation sequencing: application to the leopard cat (Prionailurus bengalensis) in Pakistan Mol Ecol 21:1951–1965. https://doi.org/10.1111/j.1365-294X.2011.05424.x
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Valentini A, Pompanon F, Taberlet P (2009) DNA barcoding for ecologists Trends Ecol Evol 24:110–117
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13:1. https://doi.org/10.1186/1471-2105-13-134
Acknowledgements
We thank CENAP (Centro Nacional de Pesquisa e Conservação de Mamíferos Carnívoros) for providing some tissue samples used in the control tests. We also thank IDEA WILD for the Geneious software license and financial support for DNA sequencing service, as well as Neotropical Grassland Conservancy (NGC) for financial support for field expeditions and samples collection. PMGJ and KGRC are thankful to Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq - 308385/2014-4, 140689/2013-3, respectively), BHS to Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP - 2013/24453-4) and DVB to Fundação Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES - 23038.006945/2011-74) for financial support. The authors are also thankful for the constructive comments of anonymous reviewer which improved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rodríguez-Castro, K.G., Saranholi, B.H., Bataglia, L. et al. Molecular species identification of scat samples of South American felids and canids. Conservation Genet Resour 12, 61–66 (2020). https://doi.org/10.1007/s12686-018-1048-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12686-018-1048-6