Abstract
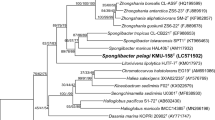
A Gram-stain-negative, rod-shaped, obligately aerobic, nonflagellated, and chemoheterotrophic bacterium, designated IMCC3088T, was isolated from coastal seawater of the Yellow Sea. The 16S rRNA gene sequence analysis indicated that this strain belonged to the family Halieaceae which shared the highest sequence similarities with Luminiphilus syltensis NOR5-1BT (94.5%) and Halioglobus pacificus S1-72T (94.5%), followed by 92.3–94.3% sequence similarities with other species within the aforementioned family. Phylogenetic analyses demonstrated that strain IMCC3088T was robustly clustered with Luminiphilus syltensis NOR5-1BT within the family Halieaceae. However, average amino acid identity (AAI), percentages of conserved proteins (POCP), average nucleotide identity (ANI), and alignment fraction (AF) between strain IMCC3088T and Luminiphilus syltensis NOR5-1BT were 54.5%, 47.7%, 68.0%, and 16.5%, respectively, suggesting that they belonged to different genera. Whole-genome sequencing of strain IMCC3088T revealed a 3.1 Mbp genome size with a DNA G + C content of 51.7 mol%. The genome encoded diverse metabolic pathways including sulfur oxidation, phenol degradation, and proteorhodopsin phototrophy. Mono-unsaturated fatty acids were found to be the predominant cellular fatty acid components in the strain. Phosphatidylethanolamine, phosphatidylglycerol, and diphosphatidylglycerol were the primarily identified polar lipids, and ubiquinone-8 was identified as a major respiratory quinone. The taxonomic data collected herein suggested that strain IMCC3088T represented a novel genus and species of the family Halieaceae, for which the name Aequoribacter fuscus gen. nov., sp. nov. is proposed with the type strain (= KACC 15529T = NBRC 108213T).
Similar content being viewed by others
References
Barco, R.A., Garrity, G.M., Scott, J.J., Amend, J.P., Nealson, K.H., and Emerson, D. 2020. A genus definition for Bacteria and Archaea based on a standard genome relatedness index. mBio 11, e02475–19.
Chang, Y.Q., Meng, X., Du, Z.Z., and Du, Z.J. 2019. Kineobactrum sediminis gen. nov., sp. nov., isolated from marine sediment. Int. J. Syst. Evol. Microbiol., 69, 2395–2400.
Chen, I.A., Chu, K., Palaniappan, K., Pillay, M., Ratner, A., Huang, J., Huntemann, M., Varghese, N., White, J.R., Seshadri, R., et al. 2019. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 47, D666–D677.
Cho, J.C. and Giovannoni, S.J. 2003. Parvularcula bermudensis gen. nov., sp. nov., a marine bacterium that forms a deep branch in the α-Proteobacteria. Int. J. Syst. Evol. Microbiol., 53, 1031–1036.
Cho, J.C. and Giovannoni, S.J. 2004. Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl. Environ. Microbiol. 70, 432–440.
Cho, J.C., Stapels, M.D., Morris, R.M., Vergin, K.L., Schwalbach, M.S., Givan, S.A., Barofsky, D.F., and Giovannoni, S.J. 2007. Polyphyletic photosynthetic reaction centre genes in oligotrophic marine Gammaproteobacteria. Environ. Microbiol. 9, 1456–1463.
Collins, M.D. and Jones, D. 1981. A note on the separation of natural mixtures of bacterial ubiquinones using reverse-phase partition thin-layer chromatography and high performance liquid chromatography. J. Appl. Bacteriol. 51, 129–134.
Csotonyi, J.T., Stackebrandt, E., Swiderski, J., Schumann, P., and Yurkov, V. 2011. Chromocurvus halotolerans gen. nov., sp. nov., a gammaproteobacterial obligately aerobic anoxygenic phototroph, isolated from a Canadian hypersaline spring. Arch. Microbiol., 193, 573–582.
Eilers, H., Pernthaler, J., Glöckner, F.O., and Amann, R. 2000. Culturability and in situ abundance of pelagic bacteria from the North Sea. Appl. Environ. Microbiol. 66, 3044–3051.
Emms, D.M. and Kelly, S. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 238.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Fuchs, B.M., Spring, S., Teeling, H., Quast, C., Wulf, J., Schattenhofer, M., Yan, S., Ferriera, S., Johnson, J., Glöckner, F.O., et al. 2007. Characterization of a marine gammaproteobacterium capable of aerobic anoxygenic photosynthesis. Proc. Natl. Acad. Sci. USA 104, 2891–2896.
Galperin, M.Y., Makarova, K.S., Wolf, Y.I., and Koonin, E.V. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269.
Giovannoni, S.J., Bibbs, L., Cho, J.C., Stapels, M.D., Desiderio, R., Vergin, K.L., Rappé, M.S., Laney, S., Wilhelm, L.J., Tripp, H.J., et al. 2005. Proteorhodopsin in the ubiquitous marine bacterium SAR11. Nature 438, 82–85.
Gosink, J.J., Woese, C.R., and Staley, J.T. 1998. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteroides group and reclassification of ‘Flectobacillus glomeratus’ as Polaribacter glomeratus comb. nov. Int. J. Syst. Evol. Microbiol., 48, 223–235.
Han, J.R., Ye, M.Q., Wang, C., and Du, Z.J. 2019. Halioglobus sediminis sp. nov. isolated from coastal sediment. Int. J. Syst. Evol. Microbiol., 69, 1601–1605.
Hunt, M., Silva, N.D., Otto, T.D., Parkhill, J., Keane, J.A., and Harris, S.R. 2015. Circlator: automated circularization of genome assemblies using long sequencing reads. Genome Biol. 16, 294.
Jang, Y., Oh, H.M., Kang, I., Lee, K., Yang, S.J., and Cho, J.C. 2011. Genome sequence of strain IMCC3088, a proteorhodopsin-containing marine bacterium belonging to the OM60/NOR5 clade. J. Bacteriol. 193, 3415–3416.
Jung, H.S., Jeong, S.E., Kim, K.H., and Jeon, C.O. 2017. Parahaliea aestuarii sp. nov. isolated from the Asan Bay estuary. Int. J. Syst. Evol. Microbiol. 67, 1431–1435.
Kanehisa, M., Sato, Y., and Morishima, K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731.
Kang, J.W., Yang, H.G., Choi, S., Kim, Y.J., and Lee, S.D. 2020. Seongchinamella unica gen. nov., sp. nov., isolated from a tidal mudflat of beach, and transfer of Halioglobus sediminis to Seongchina mella sediminis comb. nov. and Halioglobus lutimaris to Pseudohalioglobus gen. nov. as Pseudohalioglobus lutimaris comb. nov. Int. J. Syst. Evol. Microbiol., 70, 2194–2203.
Konkit, M., Kim, J.H., and Kim, W. 2016. Marimicrobium arenosum gen. nov., sp. nov., a moderately halophilic bacterium isolated from sea sand. Int. J. Syst. Evol. Microbiol., 66, 856–861.
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. 2018. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549.
Lee, J., Kwon, K.K., Lim, S.I., Song, J., Choi, A.R., Yang, S.H., Jung, K.H., Lee, J.H., Kang, S.G., Oh, H.M., et al. 2019. Isolation, cultivation, and genome analysis of proteorhodopsin-containing SAR116-clade strain Candidatus Puniceispirillum marinum IMCC1322. J. Microbiol. 57, 676–687.
Li, S.H., Song, J., Lim, Y., Joung, Y., Kang, I., and Cho, J.C. 2020. Halioglobus maricola sp. nov. disolated from coastal seawater. Int. J. Syst. Evol. Microbiol.. {b70}, 1868–1875.
Lin, C.Y., Zhang, X.Y., Liu, A., Liu, C., Song, X.Y., Su, H.N., Qin, Q.L., Xie, B.B., Zhang, Y.Z., and Chen, X.L. 2015. Haliea atlantica sp. nov., isolated from seawater, transfer of Haliea mediterranea to Parahaliea gen. nov. as Parahaliea mediterranea comb. nov. and emended description of the genus Haliea. Int. J. Syst. Evol. Microbiol., 65, 3413–3418.
Liu, Y., Du, J., Zhang, J., Lai, Q., Shao, Z., and Zhu, H. 2020. Parahaliea maris sp. nov. isolated from surface seawater and emended description of the genus Parahaliea. J. Microbiol 58, 92–98.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, Buchner, A., Lai, T., Steppi, S., Jobb, G., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Markowitz, V.M., Mavromatis, K., Ivanova, N.N., Chen, I.M., Chu, K., and Kyrpides, N.C. 2009. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics 25, 2271–2278.
Minnikin, D.E., O’Donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Na, S.I., Kim, Y.O., Yoon, S.H., Ha, S.M., Baek, I., and Chun, J. 2018. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 280–285.
Park, S., Yoshizawa, S., Inomata, K., Kogure, K., and Yokota, A. 2012. Halioglobus japonicus gen. nov., sp. nov. and Halioglobus pacificus sp. nov., members of the class Gammaproteobacteria isolated from seawater. Int. J. Syst. Evol. Microbiol., 62, 1784–1789.
Pruesse, E., Peplies, J., and Glöckner, F.O. 2012. SINA: accurate highthroughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829.
Qin, Q.L., Xie, B.B., Zhang, X.X., Chen, X.L., Zhou, B.C., Zhou, J., Oren, A., and Zhang, Y.Z. 2014. A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215.
Rodriguez-R, L.M. and Konstantinidis, K.T. 2014. Bypassing cultivation to identify bacterial species. Microbe 9, 111–118.
Rodriguez-R, L.M. and Konstantinidis, K.T. 2016. The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 4, e1900v1.
Ruff, S.E, Probandt, D., Zinkann, A.C., Iversen, M.H., Klaas, C., Würzberg, L., Krombholz, N., Wolf-Gladrow, D., Amann, R., and Knittel, K. 2014. Indications for algae-degrading benthic microbial communities in deep-sea sediments along the Antarctic Polar Front. Deep Sea Res. Part II Top. Stud. Oceanogr., 108, 6–16.
Rzhetsky, A. and Nei, M. 1993. Theoretical foundation of the minimum-evolution method of phylogenetic inference. Mol. Biol. Evol. 10, 1073–1095.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark, Delaware, USA.
Seemann, T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069.
Shi, M.J., Wang, C., Wang, X.T., and Du, Z.J. 2018. Halioglobus lutimaris sp. nov., isolated from coastal sediment. Int. J. Syst. Evol. Microbiol., 68, 876–880.
Spring, S., Lünsdorf, H., Fuchs, B.M., and Tindall, B.J. 2009. The photosynthetic apparatus and its regulation in the aerobic gammaproteobacterium Congregibacter litoralis gen. nov., sp. nov. PLoS One, 4, e4866.
Spring, S., Riedel, T., Spröer, C., Yan, S., Harder, J., and Fuchs, B.M. 2013. Taxonomy and evolution of bacteriochlorophyll a-containing members of the OM60/NOR5 clade of marine gammaproteobacteria: description of Luminiphilus syltensis gen. nov., sp. nov., reclassification of Haliea rubra as Pseudohaliea rubra gen. nov., comb. nov., and emendation of Chromatocurvus halotolerans. BMC Microbiol., 13, 118.
Spring, S., Scheuner, C., Göker, M., and Klenk, H.P. 2015. A taxonomic framework for emerging groups of ecologically important marine gammaproteobacteria based on the reconstruction of evolutionary relationships using genome-scale data. Front. Microbiol. 6, 281.
Suzuki, T., Yazawa, T., Morishita, N., Maruyama, A., and Fuse, H. 2019. Genetic and physiological characteristics of a novel marine propylene-assimilating Halieaceae bacterium isolated from seawater and the diversity of its alkene and epoxide metabolism genes. Microbes Environ. 34, 33–42.
Urios, L., Intertaglia, L., Lesongeur, F., and Lebaron, P. 2008. Haliea salexigens gen. nov., sp. nov., a member of the Gammaproteobacteria from the Mediterranean Sea. Int. J. Syst. Evol. Microbiol., 58, 1233–1237.
Urios, L., Intertaglia, L., Lesongeur, F., and Lebaron, P. 2009. Haliea rubra sp. nov. a member of the Gammaproteobacteria from the Mediterranean Sea. Int. J. Syst. Evol. Microbiol 59, 1188–1192.
Varghese, N.J., Mukherjee, S., Ivanova, N., Konstantinidis, K.T., Mavrommatis, K., Kyrpides, N.C., and Pati, A. 2015. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 43, 6761–6771.
Weisburg, W.G., Barns, S.M., Pelletier, D.A., and Lane, D.J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Yan, S., Fuchs, B.M., Lenk, S., Harder, J., Wulf, J., Jiao, N.Z., and Amann, R. 2009. Biogeography and phylogeny of the NOR5/OM60 clade of Gammaproteobacteria. Syst. Appl. Microbiol. 32, 124–139.
Yang, Q., Jiang, Z., Zhou, X., Zhang, R., Xie, Z., Zhang, S., Wu, Y., Ge, Y., and Zhang, X. 2020. Haliea alexandrii sp. nov. isolated from phycosphere microbiota of the toxin-producing dinoflagellate Alexandrium catenella. Int. J. Syst. Evol. Microbiol. 70, 1133–1138.
Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F.O., Ludwig, W., Schleifer, K.H., Whitman, W.B., Euzéby, J., Amann, R., and Rosselló-Móra, R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat. Rev. Microbiol. 12, 635–645.
Yoon, S.H., Ha, S.M., Kwon, S., Lim, J., Kim, Y., Seo, H., and Chun, J. 2017. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617.
Acknowledgments
This study was supported by the Collaborative Genome Program of the Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) (No. 20180430 to J-CC) and also by National Research Foundation of Korea (NRF) grants (NRF-2019-R1A2B5B02070538 to J-CC; NRF-2019R1I1A1A01063401 to IK).
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Li, SH., Song, J., Kang, I. et al. Aequoribacter fuscus gen. nov., sp. nov., a new member of the family Halieaceae, isolated from coastal seawater. J Microbiol. 58, 463–471 (2020). https://doi.org/10.1007/s12275-020-0206-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-020-0206-1