Abstract
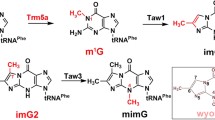
Bacterial tRNA (guanine37-N1)-methyltransferase (TrmD) plays important roles in translation, making it an important target for the development of new antibacterial compounds. TrmD comprises two domains with the N-terminal domain binding to the S-adenosyl-l-methionine (SAM) cofactor and the C-terminal domain critical for tRNA binding. Bacterial TrmD is functional as a dimer. Here we report the backbone NMR resonance assignments for the full length TrmD protein of Pseudomonas aeruginosa. Most resonances were assigned and the secondary structure for each amino acid was determined according to the assigned backbone resonances. The availability of the assignment will be valuable for exploring molecular interactions of TrmD with ligands, inhibitors and tRNA.



Similar content being viewed by others
References
Bjork G, Wikstrom P, Bystrom A (1989) Prevention of translational frameshifting by the modified nucleoside 1-methylguanosine. Science 244:986–989
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Gayen S, Li Q, Kang C (2012) The solution structure of the S4-S5 linker of the hERG potassium channel. J Pept Sci 18:140–145
Goto-Ito S, Ito T, Kuratani M, Bessho Y, Yokoyama S (2009) Tertiary structure checkpoint at anticodon loop modification in tRNA functional maturation. Nat Struct Mol Biol 16:1109–1115
Goto-Ito S, Ito T, Yokoyama S (2017) Trm5 and TrmD: two enzymes from distinct origins catalyze the identical tRNA modification, m(1)G37. Biomolecules 7:32
Hill PJ, Abibi A, Albert R, Andrews B, Gagnon MM, Gao N, Grebe T, Hajec LI, Huang J, Livchak S, Lahiri SD, McKinney DC, Thresher J, Wang H, Olivier N, Buurman ET (2013) Selective Inhibitors of bacterial t-RNA-(N1G37) methyltransferase (TrmD) that demonstrate novel ordering of the lid domain. J Med Chem 56:7278–7288
Holmes WM, Andraos-Selim C, Redlak M (1995) tRNA-m1G methyltransferase interactions: touching bases with structure. Biochimie 77:62–65
Ito T, Masuda I, Yoshida K-I, Goto-Ito S, Sekine S-I, Suh SW, Hou Y-M, Yokoyama S (2015) Structural basis for methyl-donor-dependent and sequence-specific binding to tRNA substrates by knotted methyltransferase TrmD. Proc Natl Acad Sci 112:E4197–E4205
Johnson BA (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol 278:313–352
Juhling F, Morl M, Hartmann RK, Sprinzl M, Stadler PF, Putz J (2009) tRNAdb 2009: compilation of tRNA sequences and tRNA genes. Nucleic Acids Res 37:D159–D162
Kim YM, Li Q, Ng HQ, Yoon HS, Kang C (2013) 1H, 13C and 15N chemical shift assignments for the N-terminal PAS domain of the KCNH channel from Zebrafish. Biomol NMR Assign 8:165–168
Li Q, Raida M, Kang C (2010) 1H, 13C and 15N chemical shift assignments for the N-terminal domain of the voltage-gated potassium channel-hERG. Biomol NMR Assign 4:211–213
Li Y, Wong YL, Lee MY, Ng HQ, Kang C (2016) Backbone assignment of the N-terminal 24-kDa fragment of Escherichia coli topoisomerase IV ParE subunit. Biomol NMR Assign 10:135–138
Li Y, Zhong W, Koay AZ, Ng HQ, Koh-Stenta X, Nah Q, Lim SH, Larsson A, Lescar J, Hill J, Dedon PC, Kang C (2018) Backbone resonance assignment for the N-terminal region of bacterial tRNA-(N1G37) methyltransferase. Biomol NMR Assign 13:49–53
O’Dwyer K, Watts JM, Biswas S, Ambrad J, Barber M, Brulé H, Petit C, Holmes DJ, Zalacain M, Holmes WM (2004) Characterization of Streptococcus pneumoniae TrmD, a tRNA methyltransferase essential for growth. J Bacteriol 186:2346–2354
Persson BC, Bylund GO, Berg DE, Wikstrom PM (1995) Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J Bacteriol 177:5554–5560
Pervushin K, Ono A, Fernandez C, Szyperski T, Kainosho M, Wuthrich K (1998) NMR scalar couplings across Watson–Crick base pair hydrogen bonds in DNA observed by transverse relaxation-optimized spectroscopy. Proc Natl Acad Sci USA 95:14147–14151
Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K (1998) TROSY in triple-resonance experiments: new perspectives for sequential NMR assignment of large proteins. Proc Natl Acad Sci USA 95:13585–13590
Shen Y, Bax A (2013) Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR 56:227–241
Shen Y, Delaglio F, Cornilescu G, Bax A (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J Biomol NMR 44:213–223
Thanassi JA, Hartman-Neumann SL, Dougherty TJ, Dougherty BA, Pucci MJ (2002) Identification of 113 conserved essential genes using a high-throughput gene disruption system in Streptococcus pneumoniae. Nucleic Acids Res 30:3152–3162
Thomas SR, Keller CA, Szyk A, Cannon JR, LaRonde-LeBlanc NA (2011) Structural insight into the functional mechanism of Nep1/Emg1 N1-specific pseudouridine methyltransferase in ribosome biogenesis. Nucleic Acids Res 39:2445–2457
Zhang Z, Li Y, Loh YR, Phoo WW, Hung AW, Kang C, Luo D (2016) Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 354:1597–1600
Zhong W, Koay A, Ngo A, Li Y, Nah Q, Wong YH, Chionh YH, Ng HQ, Koh-Stenta X, Poulsen A, Foo K, McBee M, Choong ML, El Sahili A, Kang C, Matter A, Lescar J, Hill J, Dedon P (2019) Targeting the bacterial epitranscriptome for antibiotic development: discovery of novel tRNA-(N(1)G37) methyltransferase (TrmD) inhibitors. ACS Infect Dis 3:326–335
Acknowledgements
CK appreciates the support from NMRC OF-IRG Grant (NMRC/OFIRG/0051/2017) and A*STAR JCO Grant (1431AFG102/1331A028). This work is also supported by National Research Foundation of Singapore through the Singapore-MIT-Alliance for Research and Technology (SMART) Infectious Disease and Antimicrobial Resistance Interdisciplinary Research Groups. WZ was supported by a SMART Scholar Fellowship. We also thank Prof Ho Sup Yoon and Dr. Hong Ye from Nanyang Technological University for the NMR experiments. The authors appreciate the valuable discussion from the team members at EDDC, A*STAR.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, Y., Zhong, W., Koay, A.Z. et al. Backbone resonance assignment for the full length tRNA-(N1G37) methyltransferase of Pseudomonas aeruginosa. Biomol NMR Assign 13, 327–332 (2019). https://doi.org/10.1007/s12104-019-09900-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12104-019-09900-2