Abstract
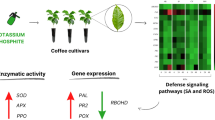
Coffea arabica is the most economically important coffee species worldwide. However, its production is severely limited by diseases such as rust. The mechanisms underlying constitutive defense responses in coffee are still poorly understood, compared with induced defense mechanisms. We aimed to characterize constitutive defense responses of thirteen cultivars of C. arabica. Cultivars were classified under field conditions according to the level of resistance to rust: resistant (R), moderately resistant (MR), and susceptible (S). Based on this classification, the stability of eight reference genes (RGs) was evaluated. The most stable RGs were EF1α, APT1, and 24S. We also evaluated the expression of CaWRKY1, CaPAL1, CaCAD1, and CaPOX1, and activities of PAL, CAD, and POX, which are involved in lignin biosynthesis, and leaf content of total phenolic compounds and lignin. Gene expression and enzymatic activity were not correlated with defense metabolites in the R cultivar group but showed a negative correlation with phenolic compounds in MR cultivars. Cultivar S showed positive correlations of gene expression and enzyme activity with phenolic compounds. These results may assist coffee breeding programs regarding selection of genotypes and in optimization of rust resistance.





Similar content being viewed by others
References
International Coffee Organization. (2020). Crop year production by country. Retrieved June 11, 2020, from http://www.ico.org/trade_statistics.asp?section=statistics
Avelino, J., Cristancho, M., Georgiou, S., Imbach, P., Aguilar, L., Bornemann, G., et al. (2015). The coffee rust crises in Colombia and Central America (2008–2013): impacts, plausible causes and proposed solutions. Food Security, 7(2), 303–321.
Capucho, A. S., Zambolim, L., Lopes, U. N., & Milagres, N. S. (2013). Chemical control of coffee leaf rust in Coffea canephora cv. conilon. Australasian Plant Pathology, 42(6), 667–673.
Talhinhas, P., Batista, D., Diniz, I., Vieira, A., Silva, D. N., Loureiro, A., et al. (2017). The coffee leaf rust pathogen Hemileia vastatrix: One and a half centuries around the tropics. Molecular Plant Pathology, 18(8), 1039–1051.
Capucho, A. S., Zambolim, E. M., Freitas, R. L., Haddad, F., Caixeta, E. T., & Zambolim, L. (2012). Identification of race XXXIII of Hemileia vastatrix on Coffea arabica Catimor derivatives in Brazil. Australasian Plant Disease Notes, 7(1), 189–191.
Zambolim, L. (2016). Current status and management of coffee leaf rust in Brazil. Tropical Plant Pathology, 41(1), 1–8.
Alkimim, E. R., Caixeta, E. T., Sousa, T. V., Pereira, A. A., de Oliveira, A. C. B., Zambolim, L., & Sakiyama, N. S. (2017). Marker-assisted selection provides arabica coffee with genes from other Coffea species targeting on multiple resistance to rust and coffee berry disease. Molecular Breeding, 37(1), 10.
Cabral, P. G. C., Maciel-Zambolim, E., Oliveira, S. A. S., Caixeta, E. T., & Zambolim, L. (2016). Genetic diversity and structure of Hemileia vastatrix populations on Coffea spp. Plant Pathology, 65(2), 196–204.
Reis, E. A. C., Freitas, T., Mendes, A. N. G., Rezende, T. T., & Carvalho, J. P. F. (2018). Characterization of coffee cultivars leaf rust-resistant subjected to framework pruning. Coffee Science, 13(1), 8.
de Carvalho, A. M., Mendes, A. N. G., Rezende, F. V., Botelho, C. E., Carvalho, G. R., & Ferreira, A. D. (2016). Selection of coffee progenies of catucaí group. Coffee Science, 11(2), 244–254.
De Carvalho, A. M., De Abreu Cardoso, D., Carvalho, G. R., De Carvalho, V. L., Pereira, A. A., Ferreira, A. D., & Carneiro, L. F. (2017). Behavior of coffee cultivars under the incidence of diseases of rust and gray leaf spot in two cultivation environments. Coffee Science, 12(1), 100–107.
Dias, R. A., Ribeiro, M. R., de Carvalho, A. M., Botelho, C. E., Mendes, A. N. G., Ferreira, A. D., & Fernandes, F. C. (2019). Selection of coffee progenies for resistance to leaf rust and favorable agronomic traits. Coffee Science, 14(2), 10.
Kushalappa, A. C., & Gunnaiah, R. (2013). Metabolo-proteomics to discover plant biotic stress resistance genes. Trends in Plant Science, 18(9), 522–531.
Caicedo, B. L. C., Cortina Guerrero, H. A., Roux, J., & Wingfield, M. J. (2013). New coffee (Coffea arabica) genotypes derived from Coffea canephora exhibiting high levels of resistance to leaf rust and Ceratocystis canker. Tropical Plant Pathology, 38(6), 485–494.
Herrera, J. C. P., Alvarado, A. G., Cortina, G. H. A., Combes, M. C., Romero, G. G., & Lashermes, P. (2009). Genetic analysis of partial resistance to coffee leaf rust (Hemileia vastatrix Berk & Br.) introgressed into the cultivated Coffea arabica L. from the diploid C. canephora species. Euphytica, 167(1), 57–67.
Ramiro, D., Jalloul, A., Petitot, A. S., Grossi de Sá, M. F., Maluf, M. P., & Fernandez, D. (2010). Identification of coffee WRKY transcription factor genes and expression profiling in resistance responses to pathogens. Tree Genetics and Genomes, 6(5), 767–781.
Kushalappa, A. C., Yogendra, K. N., & Karre, S. (2016). Plant innate immune response: Qualitative and quantitative resistance. Critical Reviews in Plant Sciences, 35(1), 38–55.
Liu, Q., Luo, L., & Zheng, L. (2018). Lignins: Biosynthesis and biological functions in plants. International Journal of Molecular Sciences, 19(2), 335.
Malinovsky, F. G., Fangel, J. U., & Willats, W. G. T. (2014). The role of the cell wall in plant immunity. Frontiers in Plant Science, 5, 1–12.
Wang, H., Avci, U., Nakashima, J., Hahn, M. G., Chen, F., & Dixon, R. A. (2010). Mutation of WRKY transcription factors initiates pith secondary wall formation and increases stem biomass in dicotyledonous plants. Proceedings of the National Academy of Sciences of the United States of America, 107(51), 22338–22343.
Jiang, J., Ma, S., Ye, N., Jiang, M., Cao, J., & Zhang, J. (2017). WRKY transcription factors in plant responses to stresses. Journal of Integrative Plant Biology, 59(2), 86–101.
Miedes, E., Vanholme, R., Boerjan, W., & Molina, A. (2014). The role of the secondary cell wall in plant resistance to pathogens. Frontiers in Plant Science, 5, 1–13.
Zhang, X., & Liu, C. J. (2015). Multifaceted regulations of gateway enzyme phenylalanine ammonia-lyase in the biosynthesis of phenylpropanoids. Molecular Plant, 8(1), 17–27.
Weng, J. K., & Chapple, C. (2010). The origin and evolution of lignin biosynthesis. New Phytologist, 187(2), 273–285.
Frei, M. (2013). Lignin: Characterization of a multifaceted crop component. The Scientific World Journal, 2013, 1–25.
Azinheira, H. G., do Silva, M. C., Talhinhas, P., Medeira, C., Maia, I., Petitot, A. S., & Fernandez, D. (2010). Non-host resistance responses of Arabidopsis thaliana to the coffee leaf rust fungus (Hemileia vastatrix). Botany, 88(7), 621–629.
Barka, G. D., Caixeta, E. T., de Almeida, R. F., Alvarenga, S. M., & Zambolim, L. (2017). Differential expression of molecular rust resistance components have distinctive profiles in Coffea arabica - Hemileia vastatrix interactions. European Journal of Plant Pathology, 149(3), 543–561.
Diniz, I., Talhinhas, P., Azinheira, H. G., Várzea, V., Medeira, C., Maia, I., et al. (2012). Cellular and molecular analyses of coffee resistance to Hemileia vastatrix and nonhost resistance to Uromyces vignae in the resistance-donor genotype HDT832/2. European Journal of Plant Pathology, 133(1), 141–157.
Guerra-Guimarães, L., Tenente, R., Pinheiro, C., Chaves, I., Silva, M. C., Cardoso, F. M. H., et al. (2015). Proteomic analysis of apoplastic fluid of Coffea arabica leaves highlights novel biomarkers for resistance against Hemileia vastatrix. Frontiers in Plant Science, 6, 1–16.
Guzzo, S. D., Harakava, R., & Tsai, S. M. (2009). Identification of coffee genes expressed during systemic acquired resistance and incompatible interaction with Hemileia vastatrix. Journal of Phytopathology, 157(10), 625–638.
Monteiro, A. C. A., de Resende, M. L. V., Valente, T. C. T., Ribeiro Junior, P. M., Pereira, V. F., da Costa, J. R., & da Silva, J. A. G. (2016). Manganese phosphite in coffee defence against Hemileia vastatrix, the coffee rust fungus: Biochemical and molecular analyses. Journal of Phytopathology, 164(11–12), 1043–1053.
Petitot, A. S., Lecouls, A. C., & Fernandez, D. (2008). Sub-genomic origin and regulation patterns of a duplicated WRKY gene in the allotetraploid species Coffea arabica. Tree Genetics and Genomes, 4(3), 379–390.
Feussner, I., & Polle, A. (2015). What the transcriptome does not tell—proteomics and metabolomics are closer to the plants’ patho-phenotype. Current Opinion in Plant Biology, 26, 26–31.
Gururani, M. A., Venkatesh, J., Upadhyaya, C. P., Nookaraju, A., Pandey, S. K., & Park, S. W. (2012). Plant disease resistance genes: Current status and future directions. Physiological and Molecular Plant Pathology, 78, 51–65.
Vieira, A., Talhinhas, P., Loureiro, A., Duplessis, S., Fernandez, D., Silva, M. C., et al. (2011). Validation of RT-qPCR reference genes for in planta expression studies in Hemileia vastatrix, the causal agent of coffee leaf rust. Fungal Biology, 115(9), 891–901.
Udvardi, M. K., Czechowski, T., & Scheible, W. R. (2008). Eleven golden rules of quantitative RT-PCR. The Plant Cell, 20(7), 1736–1737.
Kozera, B., & Rapacz, M. (2013). Reference genes in real-time PCR. Journal of Applied Genetics, 54(4), 391–406.
Budzinski, I. G. F., Moon, D. H., Morosini, J. S., Lindén, P., Bragatto, J., Moritz, T., & Labate, C. A. (2016). Integrated analysis of gene expression from carbon metabolism, proteome and metabolome, reveals altered primary metabolism in Eucalyptus grandis bark, in response to seasonal variation. BMC Plant Biology, 16(1), 149.
Dong, C., Wang, R., Zheng, X., Zheng, X., Jin, L., Wang, H., et al. (2018). Integration of transcriptome and proteome analyses reveal molecular mechanisms for formation of replant disease in Nelumbo nucifera. RSC Advances, 8(57), 32574–32587.
Capucho, A. S., Zambolim, L., Duarte, H. S. S., & Vaz, G. R. O. (2011). Development and validation of a standard area diagram set to estimate severity of leaf rust in Coffea arabica and C. canephora. Plant Pathology, 60(6), 1144–1150.
Shaner, G., & Finney, R. E. (1977). The effect of nitrogen fertilization on the expression of slow-mildewing resistance in Knox wheat. Phytopathology, 67, 1051–1056.
De Carvalho, K., Bespalhok Filho, J. C., Dos Santos, T. B., De Souza, S. G. H., Vieira, L. G. E., Pereira, L. F. P., & Domingues, D. S. (2013). Nitrogen starvation, salt and heat stress in coffee (Coffea arabica L.): Identification and validation of new genes for qPCR normalization. Molecular Biotechnology, 53(3), 315–325.
Fernandes-Brum, C. N., Garcia, B. de O., Moreira, R. O., Ságio, S. A., Barreto, H. G., Lima, A. A., et al. (2017). A panel of the most suitable reference genes for RT-qPCR expression studies of coffee: screening their stability under different conditions. Tree Genetics and Genomes. https://doi.org/10.1007/s11295-017-1213-1
Freitas, N. C., Barreto, H. G., Fernandes-Brum, C. N., Moreira, R. O., Chalfun-Junior, A., & Paiva, L. V. (2017). Validation of reference genes for qPCR analysis of Coffea arabica L. somatic embryogenesis-related tissues. Plant Cell, Tissue and Organ Culture, 128(3), 663–678.
Martins, M. Q., Fortunato, A. S., Rodrigues, W. P., Partelli, F. L., Campostrini, E., Lidon, F. C., et al. (2017). Selection and validation of reference genes for accurate RT-qPCR data normalization in Coffea spp. under a climate changes context of interacting elevated [CO2] and temperature. Frontiers in Plant Science, 8, 307.
Petitot, A. S., Barsalobres-Cavallari, C., Ramiro, D., Albuquerque Freire, E., Etienne, H., & Fernandez, D. (2013). Promoter analysis of the WRKY transcription factors CaWRKY1a and CaWRKY1b homoeologous genes in coffee Coffea arabica. Plant Cell Reports, 32(8), 1263–1276.
de Andrade, C. C. L. (2015). Cercospora coffeicola: ANÁLISES MOLECULARES , BIOQUÍMICAS E PROCESSO INFECCIOSO NA INTERAÇÃO COM CAFEEIRO. PhD Tesis, Universidade Federal de Lavras, MG, Brasil.
Xie, F., Xiao, P., Chen, D., Xu, L., & Zhang, B. (2012). miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Molecular Biology, 80(1), 75–84.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., & Speleman, F. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3(7), 12.
Pfaffl, M. W., Tichopad, A., Prgomet, C., & Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnology Letters, 26(6), 509–515.
Silver, N., Best, S., Jiang, J., & Thein, S. L. (2006). Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Molecular Biology, 7, 33.
Andersen, C. L., Jensen, J. L., & Falck Ørntoft, T. (2004). Normalization of real-time quantitative reverse transcription-PCR Data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64, 5245–5250.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research, 29, 45–45.
De Andrade, C. C. L., Vicentin, R. P., Costa, J. R., Perina, F. J., De Resende, M. L. V., & Alves, E. (2016). Alterações no metabolismo antioxidante de folhas de cafeeiro infectadas por Cercospora coffeicola. Ciencia Rural, 46(10), 1764–1770.
Possa, K. F., Silva, J. A. G., Resende, M. L. V., Tenente, R., Pinheiro, C., Chaves, I., et al. (2020). Primary metabolism is distinctly modulated by plant resistance inducers in Coffea arabica leaves infected by Hemileia vastatrix. Frontiers in Plant Science, 11, 309.
Biemelt, S., Keetman, U., & Albrecht, G. (1998). Re-aeration following hypoxia or anoxia leads to activation of the antioxidative defense system in roots of wheat seedlings. Plant Physiology, 116, 651–658.
Zucker, M. (1965). Induction of phenylalanine deaminase by light and its relation to chlorogenic acid synthesis in potato tuber tissue. Plant Physiology, 40, 779–784.
Ma, L., He, J., Liu, H., & Zhou, H. (2017). The phenylpropanoid pathway affects apple fruit resistance to Botrytis cinerea. Journal of Phytopathology, 166(3), 206–215.
Urbanek, H., Kuzniak-Gebarowska, E., & Herka, K. (1991). Elicitation of defense responses in bean-leaves by Botrytis cinerea polygalacturonase. Acta Physiologiae Plantarum, 13, 43–50.
Bradford, M. (1976). A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry, 72, 248–254.
Benitez, V., Rebollo-Hernanz, M., Hernanz, S., Chantres, S., Aguilera, Y., & Martin-Cabrejas, M. A. (2019). Coffee parchment as a new dietary fiber ingredient: Functional and physiological characterization. Food Research International, 122, 105–113.
Lima, R. B., Dos-Santos, T. B., Vieira, L. G. E., Ferrarese, M. D. L. L., Ferrarese-Filho, O., Donatti, L., et al. (2013). Heat stress causes alterations in the cell-wall polymers and anatomy of coffee leaves (Coffea arabica L.). Carbohydrate Polymers, 93(1), 135–143.
Oliveira, F. C., Srinivas, K., Helms, G. L., Isern, N. G., Cort, J. R., Gonçalves, A. R., & Ahring, B. K. (2018). Characterization of coffee (Coffea arabica) husk lignin and degradation products obtained after oxygen and alkali addition. Bioresource Technology, 257, 172–180.
Spanos, G. A., & Wrolstad, R. E. (1990). Influence of processing and storage on the phenolic composition of Thompson seedless grape juice. Journal of Agricultural and Food Chemistry, 38(7), 1565–1571.
Doster, M., & Bostock, R. (1988). Quantification of lignin formation in almond bark in response to wounding and infection by Phytophthora species. Phytopathology, 78, 473–477.
Gutierrez, L., Mauriat, M., Guénin, S., Pelloux, J., Lefebvre, J. F., Louvet, R., et al. (2008). The lack of a systematic validation of reference genes: A serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnology Journal, 6(6), 609–618.
Barsalobres-Cavallari, C. F., Severino, F. E., Maluf, M. P., & Maia, I. G. (2009). Identification of suitable internal control genes for expression studies in Coffea arabica under different experimental conditions. BMC Molecular Biology, 10(1), 1.
Cruz, F., Kalaoun, S., Nobile, P., Colombo, C., Almeida, J., Barros, L. M. G., Alves-Ferreira, M., et al. (2009). Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Molecular Breeding, 23(4), 607–616.
Figueiredo, A., Loureiro, A., Batista, D., Monteiro, F., Várzea, V., Pais, M. S., et al. (2013). Validation of reference genes for normalization of qPCR gene expression data from Coffea spp. hypocotyls inoculated with Colletotrichum kahawae. BMC Research Notes, 6(1), 388.
Goulao, L. F., Fortunato, A. S., & Ramalho, J. C. (2012). Selection of reference genes for normalizing quantitative real-time PCR gene expression data with multiple variables in Coffea spp. Plant Molecular Biology Reporter, 30(3), 741–759.
Joseph, J. T., Poolakkalody, N. J., & Shah, J. M. (2018). Plant reference genes for development and stress response studies. Journal of Biosciences, 43(1), 173–187.
Paez-Garcia, A., Sparks, J. A., de Bang, L., & Blancaflor, E. B. (2018). Plant actin cytoskeleton: New functions from old scaffold. In V. P. Sahi & F. Baluška (Eds.), Plant cell monographs (Vol. 23, pp. 103–137). Springer.
Pachauri, S., Chatterjee, S., Kumar, V., & Mukherjee, P. K. (2019). A dedicated glyceraldehyde-3-phosphate dehydrogenase is involved in the biosynthesis of volatile sesquiterpenes in Trichoderma virens—evidence for the role of a fungal GAPDH in secondary metabolism. Current Genetics, 65(1), 243–252.
Guo, L., Devaiah, S. P., Narasimhan, R., Pan, X., Zhang, Y., Zhang, W., & Wang, X. (2012). Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. The Plant Cell, 24(5), 2200–2212.
Vescovi, M., Zaffagnini, M., Festa, M., Trost, P., Lo Schiavo, F., & Costa, A. (2013). Nuclear accumulation of cytosolic glyceraldehyde-3-phosphate dehydrogenase in cadmium-stressed Arabidopsis roots. Plant Physiology, 162(1), 333–346.
Henry, E., Fung, N., Liu, J., Drakakaki, G., & Coaker, G. (2015). Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genetics, 11(4), e1005199.
Zysnarski, C. J., Lahiri, S., Javed, F. T., Martínez-Márquez, J. Y., Trowbridge, J. W., & Duncan, M. C. (2019). Adaptor protein complex-1 (AP-1) is recruited by the HEATR5 protein Laa1 and its co-factor Laa2 in yeast. Journal of Biological Chemistry, 294(4), 1410–1419.
Singh, R., Stoneham, C., Lim, C., Jia, X., Guenaga, J., Wyatt, R., et al. (2018). Phosphoserine acidic cluster motifs bind distinct basic regions on theμ subunits of clathrin adaptor protein complexes. Journal of Biological Chemistry, 293(40), 15678–15690.
Chapman, J. R., & Waldenström, J. (2015). With reference to reference genes: A systematic review of endogenous controls in gene expression studies. PLoS ONE, 10(11), e0141853.
Ganesh, D., Petitot, A. S., Silva, M. C., Alary, R., Lecouls, A. C., & Fernandez, D. (2006). Monitoring of the early molecular resistance responses of coffee (Coffea arabica L.) to the rust fungus (Hemileia vastatrix) using real-time quantitative RT-PCR. Plant Science, 170(6), 1045–1051.
Fernandez, D., Santos, P., Agostini, C., Bon, M. C., Petitot, A. S., Silva, M. C., et al. (2004). Coffee (Coffea arabica L.) genes early expressed during infection by the rust fungus (Hemileia vastatrix). Molecular Plant Pathology, 5(6), 527–536.
Ardila, H. D., Martínez, S. T., & Higuera, B. L. (2013). Levels of constitutive flavonoid biosynthetic enzymes in carnation (Dianthus caryophyllus L.) cultivars with differential response to Fusarium oxysporum f. sp. dianthi. Acta Physiologiae Plantarum, 35(4), 1233–1245.
Atanasova-Penichon, V., Pons, S., Pinson-Gadais, L., Picot, A., Marchegay, G., Bonnin-Verdal, M. N., et al. (2012). Chlorogenic acid and maize ear rot resistance: A dynamic study investigating Fusarium graminearum development, deoxynivalenol production, and phenolic acid accumulation. Molecular Plant-Microbe Interactions, 25(12), 1605–1616.
Ganapathy, G., Keerthi, D., Nair, R. A., & Pillai, P. (2016). Correlation of Phenylalanine ammonia lyase (PAL) and Tyrosine ammonia lyase (TAL) activities to phenolics and curcuminoid content in ginger and its wild congener, Zingiber zerumbet following Pythium myriotylum infection. European Journal of Plant Pathology, 145(4), 777–785.
Ramiro, D. A., Guerreiro-Filho, O., & Mazzafera, P. (2006). Phenol contents, oxidase activities, and the resistance of coffee to the leaf miner Leucoptera coffeella. Journal of Chemical Ecology, 32(9), 1977–1988.
Sade, D., Shriki, O., Cuadros-Inostroza, A., Tohge, T., Semel, Y., Haviv, Y., et al. (2015). Comparative metabolomics and transcriptomics of plant response to Tomato yellow leaf curl virus infection in resistant and susceptible tomato cultivars. Metabolomics, 11(1), 81–97.
Cipollini, D., Wang, Q., Whitehill, J. G. A., Powell, J. R., Bonello, P., & Herms, D. A. (2011). Distinguishing Defensive Characteristics in the Phloem of Ash Species Resistant and Susceptible to Emerald Ash Borer. Journal of Chemical Ecology, 37(5), 450–459.
Schrotenboer, A. C., Allen, M. S., & Malmstrom, C. M. (2011). Modification of native grasses for biofuel production may increase virus susceptibility. GCB Bioenergy, 3(5), 360–374.
Guillaumie, S., Mzid, R., Méchin, V., Léon, C., Hichri, I., Destrac-Irvine, A., et al. (2010). The grapevine transcription factor WRKY2 influences the lignin pathway and xylem development in tobacco. Plant Molecular Biology, 72(1–2), 215–234.
Naoumkina, M. A., He, X., & Dixon, R. A. (2008). Elicitor-induced transcription factors for metabolic reprogramming of secondary metabolism in Medicago truncatula. BMC Plant Biology, 8(1), 132.
Schluttenhofer, C., & Yuan, L. (2015). Regulation of specialized metabolism by WRKY transcription factors. Plant Physiology, 167(2), 295–306.
Phukan, U. J., Jeena, G. S., & Shukla, R. K. (2016). WRKY transcription factors: Molecular regulation and stress responses in plants. Frontiers in Plant Science, 7, 1–14.
Pandey, S. P., & Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiology, 150(4), 1648–1655.
Zhao, Q. (2016). Lignification: Flexibility, Biosynthesis and Regulation. Trends in Plant Science, 21(8), 713–721.
Kong, J. Q. (2015). Phenylalanine ammonia-lyase, a key component used for phenylpropanoids production by metabolic engineering. RSC Advances, 5(77), 62587–62603.
Fossdal, C. G., Nagy, N. E., Hietala, A. M., Kvaalen, H., Slimestad, R., Woodward, S., & Solheim, H. (2012). Indications of heightened constitutive or primed host response affecting the lignin pathway transcripts and phenolics in mature Norway spruce clones. Tree Physiology, 32(9), 1137–1147.
Thévenin, J., Pollet, B., Letarnec, B., Saulnier, L., Gissot, L., Maia-Grondard, A., et al. (2011). The simultaneous repression of CCR and CAD, two enzymes of the lignin biosynthetic pathway, results in sterility and dwarfism in Arabidopsis thaliana. Molecular Plant, 4(1), 70–82.
Voxeur, A., Wang, Y., & Sibout, R. (2015). Lignification: Different mechanisms for a versatile polymer. Current Opinion in Plant Biology, 23, 83–90.
Anterola, A. M., & Lewis, N. G. (2002). Trends in lignin modification: A comprehensive analysis of the effects of genetic manipulations/mutations on lignification and vascular integrity. Phytochemistry, 61, 221–294.
La Camera, S., Gouzerh, G., Dhondt, S., Hoffmann, L., Fritig, B., Legrand, M., & Heitz, T. (2004). Metabolic reprogramming in plant innate immunity: The contributions of phenylpropanoid and oxylipin pathways. Immunological Reviews, 198(1), 267–284.
Lehmann, S., Serrano, M., L’Haridon, F., Tjamos, S. E., & Metraux, J. P. (2015). Reactive oxygen species and plant resistance to fungal pathogens. Phytochemistry, 112(1), 54–62.
Shigeto, J., & Tsutsumi, Y. (2016). Diverse functions and reactions of class III peroxidases. New Phytologist, 209(4), 1395–1402.
Brown, J. K. M., & Rant, J. C. (2013). Fitness costs and trade-offs of disease resistance and their consequences for breeding arable crops. Plant Pathology, 62(S1), 83–95.
Ning, Y., Liu, W., & Wang, G. L. (2017). Balancing immunity and yield in crop plants. Trends in Plant Science, 22(12), 1069–1079.
Verne, S., Jaquish, B., White, R., Ritland, C., & Ritland, K. (2011). Global transcriptome analysis of constitutive resistance to the white pine weevil in spruce. Genome Biology and Evolution, 3(1), 851–867.
Wittstock, U., & Gershenzon, J. (2002). Constitutive plant toxins and their role in defense against herbivores and pathogens. Current Opinion in Plant Biology, 5(4), 300–307.
Xu, G., Yuan, M., Ai, C., Liu, L., Zhuang, E., Karapetyan, S., Wang, S., & Dong, X. (2017). UORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature, 545(7655), 491–494.
Llorca, C. M., Potschin, M., & Zentgraf, U. (2014). bZIPs and WRKYs: Two large transcription factor families executing two different functional strategies. Frontiers in Plant Science, 5, 169.
Bennett, R. N., & Wallsgrove, R. M. (1994). Secondary metabolites in plant defence mechanisms. New Phytologist, 127(4), 617–633.
Mishra, M. K. (2020). Advances in plant breeding strategies: nut and beverage crops, vol. 4: Genetic Resources and breeding of coffee (Coffea spp.) (pp. 488–502). Springer.
Funding
This study was funded in part by the Coordination of Improvement of Higher Education Personnel – Brazil (CAPES) – Finance Code 001, the National Institute for Coffee Science and Technology (INCT-Café), and the Research Support Foundation of the State of Minas Gerais (Fapemig).
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Mário Lúcio Vilela de Resende, Tharyn Reichel, Ana Cristina Andrade Monteiro, Natália Chagas Freitas, and Deila Magna dos Santos Botelho. The first draft of the manuscript was written by Tharyn Reichel, and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Reichel, T., de Resende, M.L.V., Monteiro, A.C.A. et al. Constitutive Defense Strategy of Coffee Under Field Conditions: A Comparative Assessment of Resistant and Susceptible Cultivars to Rust. Mol Biotechnol 64, 263–277 (2022). https://doi.org/10.1007/s12033-021-00405-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00405-9