Abstract
High rates of antimicrobial resistance (AMR) among Gram-negative pathogens (GNP) have been reported in Egypt. Antimicrobial surveillance and identifying the genetic basis of AMR provide important information to optimize patient care. In this study, we aimed to identify the beta-lactam resistance phenotypes and genotypes of multidrug-resistant (MDR) non-repetitive GNP from 3 tertiary hospitals in Egypt. WZe studied 495 non-repetitive MDR Gram-negative isolates from patients with complicated intra-abdominal infections (cIAI), complicated urinary tract infection (cUTI), and lower respiratory tract infection (LRTI), collected as part of the “Study for Monitoring Antimicrobial Resistance Trends” (SMART) conducted in 3 tertiary hospitals in Cairo, Egypt, from 2015 to 2016. Identification and susceptibility testing of GNP to antimicrobials were tested in each hospital laboratory and confirmed in a reference laboratory (International Health Management Associates (IHMA), Inc., Schaumburg, IL, USA). Molecular identification of extended-spectrum beta-lactamases (ESΒLs), AmpC, and carbapenem resistance genes was conducted in IHMA. Among the 495 MDR isolates, Klebsiella pneumoniae (K. pneumoniae) and Escherichia coli (E. coli) were the most common (52.7% and 44.2%). K. pneumoniae was most susceptible to colistin, amikacin, ertapenem, and imipenem (92.7%, 72.7%, 69.3%, and 64%, respectively). E. coli was most susceptible to colistin (100%), amikacin (94.1%), imipenem (90.4%), and ertapenem (83.6%). ESBL was detected in 96.2% and ESBL genotypes included blaCTX-M-15 (70.1%), blaTEM-OSBL (48.5%), blaSHV-OSBL (27.9%), and blaCTX-M-14 (10.7%). AmpC resistance genes were identified in 9.7% of the isolates, dominated by blaCMY-2 (5.7%). Carbapenem resistance genes were detected in 45.3% of the isolates. In K. pneumoniae, blaOXA-48 dominated (40.6%), followed by blaNDM-1 (23.7%) and blaOXA-232 (4.5%). In E. coli, the most frequent genes were blaNDM-5 (9.6%), blaOXA-181 (5.5%), blaOXA-244 (3.7%), and blaNDM-1 (3.7%). blaKPC-2 was identified in 0.4% of isolates. Notably, 32.3% of isolates carried more than one resistance gene. Our findings emphasize the continued need for molecular surveillance of MDR pathogens, implementation of strict infection control measures, and antimicrobial stewardship policies in our hospitals.
Similar content being viewed by others
Introduction
Antimicrobial resistance (AMR) among Gram-negative pathogens (GNP) increased worldwide. A high rate of AMR has been reported in Egypt since more than 20 years, among GNP causing nosocomial infections and outbreaks [1,2,3]. AMR rates have increased especially among nosocomial GNP, probably due to widespread abuse of antimicrobials including carbapenems in Egyptian hospitals and poor compliance with infection control practices [4, 5]. In small-scale studies, blaOXA, blaNDM, blaVIM, blaIMP, and blaKPC carbapenemase genes were detected in Egypt [6,7,8]. As the genetic basis of beta-lactam resistance was not yet studied at a large scale in Egypt, we aimed to molecularly characterize multidrug-resistant (MDR) GNP.
Methods
Study sites and strains
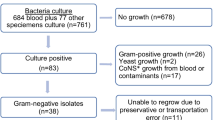
This study was conducted in 3 major tertiary care Egyptian hospitals participating in the “Study for Monitoring Antimicrobial Resistance Trends” (SMART) from 2015 to 2016. The hospitals were Ain Shams University Hospital, Ain Shams Specialized University Hospital, and Dar Al-Fouad Hospital. Isolates were collected according to SMART protocol as previously reported [9,10,11]. Briefly, the participating hospitals collected 1070 non-repetitive consecutive GNP isolates from lower respiratory tract specimens, urine and pus or abdominal fluid of hospitalized patients with lower respiratory tract infections (LRTI), complicated urinary tract infections (cUTI), and complicated intra-abdominal infections (cIAI) during the study period. Of these, we studied 495 isolates that showed phenotypic resistance to third-generation cephalosporins or carbapenems. The identification of GNP, susceptibility testing, and detection of resistance phenotypes were conducted in hospital laboratories according to the methods of the Clinical and Laboratory Standards Institute [12] and confirmed in a reference laboratory (International Health Management Associates (IHMA), Inc., Schaumburg, IL, USA), where susceptibility and extended-spectrum β-lactamase (ESBL) phenotype were determined using the CLSI broth microdilution method with custom dehydrated panels manufactured by Trek Diagnostic Systems (Thermo Scientific, Independence, OH) in 2015–2016. MIC interpretive criteria followed the 2017 M100-S27 guidelines of the CLSI [13]. EUCAST breakpoints were used only for colistin against Enterobacteriaceae, because no CLSI breakpoints exist [14].The susceptibility of all Gram-negative isolates combined was calculated using breakpoints appropriate for each species.
Genotypic identification of antimicrobial resistance genes
The molecular characterization of ESBL and carbapenemases was done using the Check-Points MDR CT103 (Check-Points Health B.V., Wageningen, The Netherlands) microarray kit, which detects most carbapenemase, ESBL, and AmpC genes: ESΒLs (class A)—blaTEM, blaSHV, blaCTX-M, blaVEB, blaPER, and blaGES; blaAmpC β-lactamase genes (class C)—blaACC, blaACT, blaCMY, blaDHA, blaFOX, blaMIR, and blaMOX; and carbapenemases (class A)—blaKPC and blaGES; (class B)—blaNDM, blaIMP, blaVIM, blaGIM, and blaSPM; and (class D)—blaOXA-48-like. Then the genes encoding ESΒL, carbapenemases, and AmpC were sequenced in their entirety in IHMA [11].
Results
Strains and phenotypic antibiotic susceptibility
The 495 MDR isolates were derived from cIAI (181, 36%), LRTI (128, 26%), and cUTI (186, 38%). Overall Klebsiella pneumoniae and Escherichia coli were the most common (52.7% and 44.2%) and were also the predominant organisms in cIAI (52.2% and 45.9%, respectively), cUTI (43% and 54.8%, respectively), and LRTI (71.1% and 26.6%, respectively) (Table 1). K. pneumoniae was most susceptible to colistin, amikacin, ertapenem, and imipenem (92.7%, 72.7%, 69.3%, and 64%, respectively). E. coli remained most susceptible to colistin (100%), amikacin (94.1%), imipenem (90.4%), and ertapenem (83.6%).
Identification of blaESBL genes of the TEM, SHV, and CTX-M types
ESBL production was detected in 96.2% of the MDR isolates (Table 2). ESBL genotypes included blaCTX-M-15 (70.1%), blaTEM-OSBL (48.5%), blaSHV-OSBL (27.9%), and blaCTX-M-14 (10.7%). The predominant ESBL gene in both E. coli and K. pneumoniae was blaCTX-M-15 (Table 3).
Identification of carbapenemase genes
Carbapenem resistance genes were detected in 45.3% of the MDR isolates. In K. pneumoniae, blaOXA-48 dominated (40.6%), followed by blaNDM-1 (23.7%) and blaOXA-232 (4.5%). In E. coli, the most frequent genes were blaNDM-5 (9.6%), blaOXA-181 (5.5%), blaOXA-244 (3.7%), and blaNDM-1 (3.7%). blaKPC-2 and blaVIM-2 were less frequently identified (Table 4).
Identification of AmpC β-lactamases resistance genes
AmpC resistance genes were identified in 9.7% of the isolates; blaCMY-2 was the most predominant one (Table 5). In 153 isolates (32.3%), coexistence of 2 or more resistance genes was detected (Table 4). The commonest combination of 2 genes was blaCTX-M-15 with blaNDM-5 (2.6%); the commonest combination of 3 genes was blaSHV-OSBL, blaCTX-M-15, and blaNDM-1 (9.2%); the commonest combination of 4 genes was blaSHV-OSBL, blaTEM-OSBL, blaCTX-M-15, and blaNDM-1 (12.4%); and the commonest combination of more than 4 genes was blaSHV-OSBL, blaTEM-OSBL, blaCTX-M-15, blaCTX-M-14, blaNDM-1, and blaOXA-48 (5.9%) (Table 6). Tables 7 and 8 show K. pneumoniae and E.coli susceptibility (%) against MDR- GNP per type of infection.
Discussion
Surveillance for AMR is essential to monitor trends, identify emerging resistance mechanisms, and support the antimicrobial stewardship programs. To our knowledge, this is the most extensive molecular study of AMR in Egypt including 495 MDR Gram-negative isolates. In all hospitals, more than half of infections were present on admission (community acquired or transferred from other hospitals; data not shown). We identified 43 resistance phenotypes distributed among isolates from the 3 hospitals. Typing of nosocomial GNP in each hospital based on the phenotypic resistance patterns showed no clonal spread except for few isolates in each hospital (data not shown).
K. pneumoniae was a common pathogen in all 3 types of infection, and most isolates were MDR. Due to its large accessory genome including plasmids and chromosomal loci, K. pneumoniae isolates may act as opportunistic pathogens. Such strains infect critically ill and immunocompromised patients mostly, whereas other strains of K. pneumoniae (hyper-virulent) may even infect healthy people in community settings. Many of the virulent strains encode carbapenemases [15]. E. coli was a frequently identified pathogen from cIAI and cUTI, which is consistent with previous reports, and was also a frequent pathogen in LRTI. E. coli pneumonia is uncommon and may result from micro-aspiration of colonized upper airway secretions in severely ill patients; hence, it is a well-known cause of nosocomial pneumonia [16]. However, E. coli pneumonia may also be community-acquired in patients who have underlying diseases such as diabetes mellitus, alcoholism, chronic obstructive pulmonary disease, and E. coli UTI [17]. Our results confirm the results of previous studies in Egypt [3, 18] that showed high rates of ESΒL and carbapenem resistance among ICU pathogens. For example, in the study of Talaat et al. [18], ESBL and carbapenem resistance were identified in 54% and 13.8% of E. coli isolates compared with 42.5% and 48.1% in K. pneumoniae isolates, respectively.
Among the many types of ESΒLs reported, blaCTX-M-15 and blaCTX-M-14 are the most commonly identified worldwide as the genes encoding CTX-M enzymes (blaCTX-M) can be horizontally mobilized by various genetic elements [19]. This reflects the situation in Egypt as well. AmpC genes were less frequently identified (9.7%), usually in combination with other resistance genes. CMY-2-like enzymes were the most predominant. Other detected genes were blaCMY-42, blaDHA, and blaACT-like. This is in keeping with recent reports on acquisition of plasmid-mediated cephalosporinase producing Enterobacteriaceae after a travel to the tropics and North Africa including Egypt [20, 21]. The high rate of ESΒL in Egypt is essentially due to inappropriate use of antimicrobials in human and animal health care. Of the patients treated in public outpatient health care facilities, 49.8% received antibiotics, and antibiotics are still sold over the counter without prescription [22]. Moreover, there is extensive use of antimicrobials for prevention and treatment of infections in veterinary care, and ESΒL and AmpC resistance mechanisms were detected in veterinary E. coli isolates in Egypt [23]. This is an urgent public health problem especially with a growing body of evidence supporting foodborne transmission of resistance, especially from poultry, as poultry meat exhibits the highest levels of contamination by MDR bacteria [24, 25].
The most important mechanism of carbapenem resistance is the production of carbapenemases; therefore, all isolates were investigated to identify the carbapenemase genes. We identified them in 45.4% of tested isolates; blaOXA followed by blaNDM genes dominated, while only 2 isolates of K. pneumoniae harbored blaKPC-2 genes. These results confirm previous small-scale reports that the blaNDM and blaOXA genes are the predominant in Egypt and Middle East [26].
The present study detected massive coexistence of different resistance genes among tested isolates. This coexistence could have contributed to the observed elevated variability in resistance phenotypes and genotypes among GNP in Egypt [15].
In conclusion, our study detected alarming rates of resistance and identified many resistance mechanisms in clinical GNP from Egyptian tertiary care hospitals. These high resistance rates highlight the importance of continuous monitoring of the resistance trends, adherence to infection control policies, and underscore urgently implementing a national antimicrobial stewardship plan in Egypt.
References
El-Kholy A, Baseem H, Hall GS, Procop GW, Longworth D (2003) Antimicrobial resistance in Cairo, Egypt 1999–2000: a survey of five hospitals. J Antimicrob Chemother 51:625–630. https://doi.org/10.1093/jac/dkg101
Moore K, Kainer M, Badrawi N, Afifi S, Wasf M, Bashir M, Jarvis W, Graham T, El Kholy A, Gipson R, Jernigan D, Mahoney F (2005) Neonatal sepsis in Egypt associated with bacterial contamination of glucose-containing intravenous fluids. Pediatr Infect Dis J24:590–594. https://doi.org/10.1097/01.inf.0000168804.09875.95
See I, Lessa F, Abo El-Ata O, Hafez S, Samy K, El-Kholy A, El Anany M, Ismail G, Kandeel A, Galal R, Ellingson K, Talaat M (2013) Incidence and pathogen distribution of healthcare-associated infections in pilot hospitals in Egypt. Infect Control Hosp Epidemiol 34(12):1281–1288. https://doi.org/10.1086/673985
Saied T, Elkholy A, Hafez SF, Basim H, Wasfy MO, El-Shoubary W, Samir A, Pimentel G, Talaat M (2011) Antimicrobial resistance in pathogens causing nosocomial bloodstream infections in university hospitals in Egypt. Am J Infect Contro l39:e61–e65. https://doi.org/10.1016/j.ajic.2011.04.009
Talaat M, Saied T, Kandeel A, Abo El-Ata G, El-Kholy A, Hafez S, Osman A, Abdel Razik M, Ismail G, El-Masry S, Galal R, Yehia M, Amira A, Calfee DP (2014) A point prevalence survey of antibiotic use in 18 hospitals in Egypt. Antibiotics 3:450–460. https://doi.org/10.3390/antibiotics3030450
Amer WH, Khalil HS, Abdel Wahab MAA (2016) Risk factors, phenotypic and genotypic characterization of carbapenem resistant Enterobacteriaceae in Tanta University Hospitals, Egypt. Int J Infect Control v12:i2. https://doi.org/10.3396/ijic.v12i2.15905
Abdulall AK, Tawfick MM, El Manakhly AR, El-KholyA (2018) Carbapenem-resistant Gram-negative bacteria associated with catheter-related bloodstream infections in three intensive care units in Egypt. Eur J Clin Microbiol Infect Dis 37(9):1647–1652. https://doi.org/10.1007/s10096-018-3294-7
El-Kholy AA, Elanany MG, Sherif MM, Gad MA (2018) High prevalence of VIM, KPC, and NDM expression among surgical site infection pathogens in patients having emergency surgery. Surgical Infections (Larchmt) 19(6):629–633
Hawser SP, Bouchillon SK, Hoban DJ, Badal RE (2009) In vitro susceptibilities of aerobic and facultative anaerobic Gram-negative bacilli from patients with intra-abdominal infections worldwide from 2005–2007: results from the SMART study. Int J Antimicrob Agents 34(6):585–588. https://doi.org/10.1016/j.ijantimicag
Hawser SP, Bouchillon SK, Lascols C, Hackel M, Hoban DJ, Badal RE, Woodford N, Livermore DM (2011) Susceptibility of Klebsiella pneumoniae isolates from intra-abdominal infections and molecular characterization of ertapenem-resistant isolates. Antimicrob. Agents Chemother 55(8):3917–3921. https://doi.org/10.1128/AAC.00070-11
Morrissey I, Hackel M, Robert Badal R (2013) A review of ten years of the study for monitoring antimicrobial resistance trends (SMART) from 2002 to 2010. Pharmaceuticals 6:1335–1346
Clinical and Laboratory Standards Institute (2015) Performance standards for antimicrobial susceptibility testing. In: Twenty-Fifth Informational Supplement M100-S25. CLSI, p 2015
Clinical and Laboratory Standards Institute (2017) Performance standards for antimicrobial susceptibility testing. In: Twenty-Fifth Informational Supplement M100-S27. CLSI, p 2017
European Committee on Antimicrobial Susceptibility Testing (EUCAST) (2017) Breakpoint tables for interpretation of MICs and zone diameters Version 7.1
Martin RM, Bachman MA (2018) Colonization, infection, and the accessory genome of Klebsiella pneumoniae. Front Cell Infect Microbiol 22(8):4. https://doi.org/10.3389/fcimb.2018.00004
Linhares I, Raposo T, Rodrigues A, Almeida A (2013) Frequency and antimicrobial resistance patterns of bacteria implicated in community urinary tract infections: a ten-year surveillance study (2000–2009). BMC Infect Dis 13:19. https://doi.org/10.1186/1471-2334-13-19
Jonas M, Cunha BA (1982) Bacteremic Escherichia coli pneumonia. Arch Intern Med 142(12):2157–2159
Talaat M, El-Shokry M, El-Kholy J, Ismail G, Kotb S, Hafez S, Attia E, Lessa F (2016) National surveillance of health care–associated infections in Egypt: developing a sustainable program in a resource-limited country. Am J Infect Control1 44(11):1296–1301. https://doi.org/10.1016/j.ajic
Zhao WH, Hu ZQ (2013) Epidemiology and genetics of CTX-M extended-spectrum beta-lactamases in Gram-negative bacteria. Crit Rev Microbiol:79–101. https://doi.org/10.3109/1040841X
von Wintersdorff CJH, Penders J, Stobberingh EE, Oude Lashof AML, Hoebe CJPA, Savelkoul PHM, Wolffs FG (2014) Rates of antimicrobial drug resistance gene acquisition after international travel, The Netherlands. Emerg Infect Dis 20:649–657. https://doi.org/10.3201/eid2004.131718
Lorme F, Maataoui N, Rondinaud E, Esposito-Farèse M, Clermont O, Ruppe E, et al. (2018) Acquisition of plasmid-mediated cephalosporinase producing Enterobacteriaceae after a travel to the tropics. PLoS One 13(12): e0206909. https://doi.https://doi.org/10.1371/journal.pone.0206909
World Health Organization (WHO) (2011) Pharmaceutical Country Profile, Egypt.http://apps.who.int/medicinedocs/documents/s19733en/s19733en.pdf
El-Shazly DA, Nasef SA, Mahmoud FF, Jonas D (2017) Expanded spectrum β-lactamase producing Escherichia coli isolated from chickens with colibacillosis in Egypt. PoultSci. 96(7):2375–2384 https://doi.org/10.3382/ps/pew493
Dutil L, Irwin R, Finley R, Ng LK, Avery B, Boerlin P, Bourgault AM, Cole L, Daignault D, Desruisseau A, Demczuk W, Hoang L, Horsman GB, Ismail J, Jamieson F, Maki A, Pacagnella A, Pillai DR (2010) Ceftiofur resistance in Salmonella enterica serovar Heidelberg from chicken meat and humans, Canada. Emerg Infect Dis 16(1):48–54. https://doi.org/10.3201/eid1601.090729
Manges AR, Johnson JR (2012) Food-borne origins of Escherichia coli causing extraintestinal infections. Clin Infect Dis 55(5):712–719. https://doi.org/10.1093/cid/cis502
Khalifa HO, Soliman AM, Ahmed AM, Shimamoto T, Hara T, Ikeda M, Kuroo Y, Kayama S, Sugai M, TShimamoto T (2017) High carbapenem resistance in clinical Gram-negative pathogens isolated in Egypt. Microb Drug Resist 23(7):838–844. https://doi.org/10.1089/mdr.2015.0339
Funding
This study was part of the “Study for Monitoring Antimicrobial Resistance Trends” (SMART), funded by Merck & Co., Inc. in 192 sites in 54 countries. Merck & Co. supported laboratory supplies in hospitals, cost of transportation of isolates to the reference laboratory, and cost of testing in the reference laboratory.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
El-Kholy, A.A., Girgis, S.A., Shetta, M.A.F. et al. Molecular characterization of multidrug-resistant Gram-negative pathogens in three tertiary hospitals in Cairo, Egypt. Eur J Clin Microbiol Infect Dis 39, 987–992 (2020). https://doi.org/10.1007/s10096-020-03812-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-020-03812-z