Abstract
Background
Infectious diseases are among the leading causes of death in many low-income countries, such as Ethiopia. Without reliable local data concerning causative pathogens and antimicrobial resistance, empiric treatment is suboptimal. The objective of this study was to characterize gram-negative bacteria (GNB) as pathogens and their resistance pattern in hospitalized patients with infections in central Ethiopia.
Methods
Patients ≥ 1 year of age with fever admitted to the Asella Referral and Teaching Hospital from April 2016 to June 2018 were included. Blood and other appropriate clinical specimens were collected and cultured on appropriate media. Antibiotic susceptibility testing (AST) was performed using the Kirby–Bauer method and VITEK® 2. Species identification and detection of resistance genes were conducted using MALDI-ToF MS (VITEK® MS) and PCR, respectively.
Results
Among the 684 study participants, 54.2% were male, and the median age was 22.0 (IQR: 14–35) years. Blood cultures were positive in 5.4% (n = 37) of cases. Among other clinical samples, 60.6% (20/33), 20.8% (5/24), and 37.5% (3/8) of swabs/pus, urine and other body fluid cultures, respectively, were positive. Among 66 pathogenic isolates, 57.6% (n = 38) were GNB, 39.4% (n = 26) were gram-positive, and 3.0% (n = 2) were Candida species. Among the isolated GNB, 42.1% (16/38) were Escherichia coli, 23.7% (9/38) Klebsiella pneumoniae and 10.5% (4/38) Pseudomonas aeruginosa.
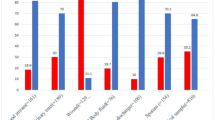
In total, 27/38 gram-negative isolates were available for further analysis. Resistance rates were as follows: ampicillin/sulbactam, 92.6% (n = 25); cefotaxime, 88.9% (n = 24); ceftazidime, 74.1% (n = 20); cefepime, 74.1% (n = 20); gentamicin, 55.6% (n = 15); piperacillin/tazobactam, 48.1% (n = 13); meropenem, 7.4% (n = 2); and amikacin, 3.7% (n = 1). The blaNDM-1 gene was detected in one K. pneumoniae and one Acinetobacter baumannii isolate, which carried an additional blaOXA-51 gene. The ESBL enzymes were detected in 81.5% (n = 22) of isolates as follows: TEM, 77.2% (n = 17); CTX-M-1 group, 68.2% (n = 15); SHV group, 27.3% (n = 6); and CTX-M-9 group, 9.1% (n = 2). Based on the in vitro antimicrobial susceptibility results, empiric treatment initiated in 13 of 18 (72.2%) patients was likely ineffective.
Conclusion
We report a high prevalence of ESBL-producing bacteria (81.5%) and carbapenem resistance (7.4%), with more than half of GNB carrying two or more ESBL enzymes resulting in suboptimal empiric antibiotic therapy. These findings indicate a need for local and national antimicrobial resistance surveillance and the strengthening of antimicrobial stewardship programs.
Similar content being viewed by others
Introduction
Infectious diseases are among the leading causes of morbidity and mortality in many low-income countries, such as Ethiopia [1]. Bacterial sepsis is a severe complication of different infections caused by gram-negative bacteria (GNB), leading to high rates of mortality. Pneumonia, bacteremia, urinary tract infection, intra-abdominal infection and skin and soft tissue infection are the major GNB sources of infections [2].
The emerging multidrug resistance (MDR) in GNB has become a serious public health problem worldwide [3]. A major driver of resistance in GNB is the horizontal transfer of mobile genetic elements carrying genes for extended-spectrum β-lactamases (ESBL) and/or carbapenemases [4]. These enzymes hydrolyze penicillins, third-generation cephalosporins (3GCs) and carbapenems. The most common ESBLs are cefotaximase (CTX-M-type), temoniera (TEM), and sulfhydryl reagent variable (SHV). Furthermore, the most prevalent carbapenemases are oxacillinases (OXA), namely, OXA-23, OXA-24/40, OXA-48, OXA-51, OXA-58, OXA-143, and OXA-235; Klebsiella pneumoniae carbapenemase (KPC); metallo-beta-lactamases (MBL), such as the New Delhi metallo-β-lactamase (NDM); imipenemase (IMP); and Verona imipenemase (VIM). These enzymes confer resistance to cephalosporins and carbapenems [5, 6].
Despite evidence for high rates of 3GC resistance among gram-negative isolates in many African countries, 3GCs are among the most commonly used antibiotics for the initial empiric treatment of severe infections and sepsis in sub-Saharan Africa (SSA). Data to estimate the mortality associated with antimicrobial resistance (AMR) in SSA are limited [7]. Implementation of antimicrobial stewardship (AMS) programs, improvement of antimicrobial susceptibility testing (AST) capacity, and provision of local and national AMR data for common bacterial pathogens should be given attention in these regions. Continued extensive empiric use of 3GC in the absence of AST might lead to a further decrease in antimicrobial activity and therefore increase the burden on the resource-limited health care systems of the countries affected [8]. This development can also be observed locally, and previous investigations at the study site show that the effectiveness of the limited choice of available antimicrobials is diminished due to the high rate of ESBL-producing GNB [9]. Due to the increase in ESBL-producing GNB-associated infections that are difficult or impossible to treat under local circumstances, there is a growing need for strategies to improve the prudent use of antibiotics in this country [10, 11].
The lack of systematically acquired local data concerning both the causative organisms and common resistance patterns likely results in ineffective empiric treatment and an unfavorable clinical outcome. Therefore, we assessed the rate and extent of drug resistance among GNB isolated from hospitalized patients with infectious diseases at a tertiary hospital in central Ethiopia and characterized the AMR genes.
Materials and methods
Study design and inclusion
This prospective operational research was conducted from April 2016 to June 2018 at the Asella Referral and Teaching Hospital (ARTH), a tertiary hospital in the town of Asella, located in the central part of Ethiopia. The bacterial cultures were performed in Ethiopia, and secondary and molecular biological investigations were performed in Germany.
During the study period, all patients ≥ 1 year of age presenting for treatment at ARTH with fever defined as body temperature > 37.5 °C according to tympanic measurement were offered inclusion in the study (Fig. 1). The eligible patients who fulfilled the inclusion criteria were identified by a trained study team. After written informed consent was obtained from the patients or legal guardians, blood cultures (BCs) were drawn from all participants. Previously initiated antibiotic treatment was not an exclusion criterion. In addition to BC, appropriate clinical samples according to the patient’s symptoms and the treating physician’s decision were collected for further microbiological investigation, according to local standard operating procedures (SOPs) and national treatment guidelines. Sociodemographic and clinical data were collected by using a standardized questionnaire.
Ethical approval and consent to participate
The appropriate ethical review boards of Arsi University (reference number A/U/H/S/C/120/6443/2017), the Oromia Regional Health Bureau (reference number BEFO/AHBTFH/1-8/2017), and Düsseldorf University Hospital (reference number 5729) approved the study. Ethical clearance for sample transportation between Ethiopia and Germany was obtained from the National Ethical Review Board of the Ministry of Science and Technology (reference number 310/204/2017). Before inclusion in the study, written informed consent to participate in the study was obtained from each participant or, in the case of minors, from their legal guardians.
Blood culture
Approximately 5 ml of blood from each child and 10–20 ml from each adult participant was collected and inoculated into an aerobic blood culture bottle. For the first 200 participants, we used in-house-produced blood culture bottles, and for the remaining participants, we used commercially available blood culture bottles (BacT/ALERT culture media bottles, bioMérieux, Marcy-l’Étoile, France). In both cases, we used the same incubator and did not find a significant difference in the positivity rates except for slightly more contaminants when using the in-house-produced blood culture bottles. The cultures were incubated at 37 °C for a maximum period of five days. After 24 h and at the end of the incubation period, Gram staining and subculturing on blood, MacConkey, and chocolate agar in a candle jar or a 5% CO2-enriched atmosphere were performed. Biochemical identification tests were subsequently performed based on the gram staining results. For GNB identification, the biochemical identification tests performed were oxidase, lactose, indole, mannitol, urease, triple sugar iron, citrate, and lysine decarboxylase, whereas for gram-positive bacteria, catalase, coagulase and hemolysis tests were used. Only one aerobic culture was performed per patient for in-house blood culture bottles, and a single pair (one aerobic and one anaerobic culture media bottle) was performed for BacT/ALERT blood culture bottles. Regular quality controls for sensitivity were performed by incubation of ATCC 25922 (Escherichia [E.] coli), ATCC 700603 (Klebsiella [K.] pneumoniae), ATCC 27853 (Pseudomonas [P.] aeruginosa), ATCC 747 (Acinetobacter [A.] baumannii) and ATCC 25923 (Staphylococcus [S.] aureus).
Urine culture
In participants with possible urinary tract infection, mid-stream urine specimens were collected using sterile urine cups, and 1 μL of urine was inoculated onto blood and MacConkey agar. We used a semiquantitative urine culture method to interpret the results for female participants. Hence, after 24 h of incubation at 37 °C, bacterial colonies were further processed if the colony count was ≥ 105 CFU/mL.
Swabs and other body fluid culture
If the patient had abscesses or any wounds, swabs or aspirates of other body fluids were collected using appropriate sterile sampling devices (swabs, sterile syringes, etc.) and cultivated on blood and chocolate agar in a 5% CO2-enriched atmosphere. The isolates were further processed following the local laboratory’s SOPs.
Antimicrobial susceptibility testing
Following the identification of GNB, Kirby–Bauer AST was performed after cultivation on Mueller–Hinton agar. The procedure and interpretation followed the European Committee on Antimicrobial Susceptibility Testing (EUCAST) version 7.1 recommendations. The microbiological results were reported to the treating physicians at ARTH as soon as possible to ensure appropriate patient care.
Confirmation of species identification and antimicrobial susceptibility test
Bacterial isolates were preserved at −80 °C in Microbank® vials (Pro-Lab Diagnostics Inc., Toronto, Canada). Subsequently, samples of all isolates were exported to Germany for confirmation of species identification and AST as well as molecular biological analysis of resistance mechanisms. Species identification was confirmed by using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry (Vitek® MS, bioMérieu), and the local AST result was confirmed by VITEK® 2 (bioMérieux). There were no significant discrepancies between the Kirby–Bauer and VITEK AST results. However, some discrepancies between the species identification according to the biochemical tests performed in Ethiopia and the MALDI-TOF results occurred. For instance, 81.5% (22/27) of GNB isolates were correctly identified by biochemical tests performed in Ethiopia compared with the MALDI-TOF results.
ESBL and carbapenemase gene detection
After identification and AST, GNB DNA was extracted by suspending a pure colony grown on MacConkey agar in 200 μL of Tris–EDTA pH 8.0 buffer. The suspension was then heated at 95 °C for 20 min, followed by centrifugation at 10,000 rpm for 10 min. Then, 150 µl of the supernatant was transferred into a new tube and stored at −20 °C until PCR testing could be performed.
All available GNB isolates were investigated by PCR (Fig. 1). We used the PCR protocols described by Strauß et al. for identification of the β-lactamase blaCTX-M, blaSHV and blaTEM genes [12] for all available GNB isolates and an in-house PCR protocol established for the detection of the carbapenemase genes balIMP-1, balVIM-1, balVIM-2, blaGIM-1, blaOXA-23, blaOXA-24/40, blaOXA-58, blaNDM-1 and blaOXA-51, as described by Wendel et al. [13, 14] (Table 1) for strains that were resistant to imipenem/meropenem according to the AST results.
Data analysis
Data were entered and analyzed using IBM SPSS Statistics for Windows version 25 (IBM Corp., Armonk, NY, USA). Fisher’s exact and Pearson’s chi-squared tests were used for the analysis of quantitative variables, and the mean, median and standard deviation were used for continuous variables. Differences were considered statistically significant at p < 0.05.
Quality control
For quality control, the control strains listed in Table 2 were used during the resistance gene analysis.
Results
In total, 684 study participants were included; 54.2% were male, and the median age was 22 years (interquartile range: 14–35 years). More than half of the participants were from rural areas, and 167 (24.4%) could not read or write (Table 3).
Bacterial cultures
The overall bacterial detection rate across all sample materials was 10.9% (83/761). In 8.7% (n = 66), the isolated bacterium was considered a pathogen, and in 2.2% (n = 17), coagulase-negative staphylococci (CoNS) were isolated and considered clinically irrelevant because of the high likelihood of contamination. The overall blood culture positivity rate was 5.4% (37/684), among which 51.4% (n = 19) were GNB. The overall detection rates in other clinical samples, namely, swabs/pus, urine, and other body fluids, were 60.6% (20/33), 20.8% (5/24), and 37.5% (3/8), respectively. Out of 12 cultured cerebrospinal fluid samples, only one revealed growth of the pathogen Neisseria meningitidis (N. meningitidis). Overall, 66 pathogenic bacteria were isolated from a total of 761 different clinical samples. Of those, 57.6% (n = 38) were GNB, 39.4% (n = 26) were gram-positive, and 3% (n = 2) were Candida species. Staphylococcus aureus and E. coli were the most prevalent isolates among the gram-positive and gram-negative isolates, respectively. Among the 38 g-negative isolates, 42.1% (n = 16) were E. coli, 23.7% (n = 9) were K. pneumoniae, and 10.5% (n = 4) were P. aeruginosa (for further details see Table 4).
As indicated in Table 5, among all clinical samples, bacterial growth was identified significantly more often among patients with skin and soft tissue infections (SSTIs) (p < 0.001). Although not statistically significant, samples from patients with a leukocyte count > 12,000 or < 4000 cells/µL tended to be more likely to reveal bacterial growth (p = 0.07). On the other hand, BCs from patients with a diagnosis of acute febrile illness of unknown source were least likely to verify bacteremia compared with other symptoms (p = 0.005). Being HIV positive had no statistically significant effect on the blood culture positivity rate (p = 0.85). There was no statistically significant difference between the mean value of C-reactive protein (CRP) in positive and negative BCs (mean CRP 71.7 mg/L vs. 64.6 mg/L; p = 0.20).
Regarding the culture positivity rate among different clinical samples, there was no significant difference between the positivity rates of gram-positive or gram-negative isolates. See Table 6 for details.
Antimicrobial susceptibility testing
In total, 27 gram-negative isolates were available for susceptibility testing with VITEK® 2. Among those, the resistance rates against commonly used antibiotics at the study site were as follows: ampicillin/sulbactam, 93.3% (n = 21); cefotaxime, 88.9% (n = 24); ceftazidime, 74.1% (n = 20); cefepime, 74.1% (n = 20); ciprofloxacin, 70.4% (n = 19); and gentamicin, 63.0% (n = 17). The resistance rates against rarely used antibiotics were 48.1% (n = 13) for piperacillin/tazobactam and 7.4% (n = 2) for meropenem. Only one case of amikacin resistance was detected among all 27 isolates (Table 7).
Resistance genes
The overall frequencies of ESBL and carbapenemase detection among the isolated GNB were 81.5% (22/27) and 7.4% (2/27), respectively. In 55.6% of cases (n = 15), more than one ESBL enzyme was detected in the isolated GNB. The different ESBL enzymes were characterized as TEM (n = 17, 77.3%), CTX-M-1 group (n = 15, 68.2%), SHV group (n = 6, 27.3%) and CTX-M-9 group (n = 2, 9.1%). Both CTX-M-1-type and SHV-type were detected in 5/6 (83.3%) of the K. pneumoniae isolates, whereas the SHV group was detected only in 1/13 of the E. coli isolates. In E. coli, only the CTX-M-1 group was common (see Table 8). Regarding carbapenemases, a single blaNDM-1 from one isolated K. pneumoniae and a combination of blaNDM-1 plus blaOXA-51 from an isolate of A. baumannii were detected.
Effectiveness of empiric antibiotic treatment
The results of the locally performed Kirby–Bauer disc diffusion test were available from 25 study participants with E. coli and K. pneumoniae isolates. Eighteen of these 25 study participants received empirically initiated antibiotic treatment at the time of sampling. The Kirby–Bauer AST revealed high levels of resistance against commonly used antibiotics, rendering 72.2% (13/18) of the initiated antibiotic treatments likely to be ineffective. In particular, 72.0% (18/25) of the isolated GNB were resistant to 3GC, 60.0% (15/25) to fluoroquinolones and 48.0% (12/25) to gentamicin. The results of the initial clinical evaluation, empiric antibiotic treatment and AST results according to the Kirby–Bauer disc diffusion test are summarized below (Table 9).
Discussion
To date, infectious diseases are one of the most common causes of morbidity and mortality in resource-limited settings, such as Ethiopia [1], but the availability of epidemiological data about causative pathogens and the distribution of AMR remains limited. In this study, we found a high rate of MDR in GNB isolated from febrile patients. As previously described in Uganda, MDR GNB is the main cause of sepsis in febrile cancer patients, with more than 50% of sepsis episodes being caused by E. coli infections [15]. Similar to these findings and to the findings of Wasihun et al. (2015) and Moges et al. (2021) from northern Ethiopia [16, 17], E. coli was the most prevalent isolated GNB in our study.
The culture positivity rate of 5.4% from BCs was low, which might partially be explained by the fact that the causative pathogen was noncultivable in a proportion of patients (hemoparasites, viruses, or noncultivable bacteria). Similar findings were reported in South Africa [18]. In patients with febrile illness with an unknown source of infection in comparison to other clinical diagnoses, the likelihood of positive BCs was lowest. The highest yield of bacterial cultures was reported from swabs in patients with SSTIs. There was a tendency toward an association between a decreased or elevated leucocyte count or an increased CRP level and blood culture positivity. However, these associations were not significant, and the predictive value of these parameters to guide blood culture diagnostics is insufficient. Thus, our finding confirms a previous report from Italy that CRP level alone is not sufficient to predict blood culture positivity [19]. In this time of widespread AMR and the associated risk of failing antibiotic therapies, blood culture diagnostics are essential to guide the management of bacterial infections. If the broad application of blood culture diagnostics is not possible due to resource limitations, the application of other parameters, such as procalcitonin or monocyte distribution width (MDW), could help to guide the rational use of BCs [20, 21].
In our study, the resistance rates of isolated GNB against commonly used antibiotics were high, severely confining the effectiveness of aminopenicillins in combination with beta-lactamase inhibitors or 3GC for empiric treatment of infections possibly caused by GNB. The lowest resistance rates were found for the meropenem and amikacin. These results are consistent with the results of other recently published data from different parts of Ethiopia, in particular from Addis Ababa [11, 22], Jimma [23] and Bahir Dar [10]. The different resistance rates to certain antibiotics reflect the frequency of antibiotic prescriptions. While aminopenicillins and 3GCs are applied very frequently, carbapenems are hardly used due to their high cost and limited availability. Of note, while the resistance rate for amikacin was very low, many of the isolated GNB were resistant to gentamicin. This difference might be explained by the frequent application of gentamicin at the study center. In contrast, amikacin is almost never applied [24, 25].
Our data revealed overall frequencies of ESBL and carbapenemase production of 81.5% and 7.4% among the isolated GNB, respectively. Of note, all isolates of K. pneumoniae were ESBL-positive. These findings are consistent with recently published data from other parts of Ethiopia, where K. pneumoniae has also been shown to be the most common ESBL-expressing pathogen, followed by E. coli [10, 11, 22].
Regarding the characterization of ESBL enzymes among the gram-negative isolates in our study, TEM-type and CTX-M-1-type were most common, followed by SHV-type and, least frequently, CTX-M-9-type. CTX-M-2 and CTX-M-8/25 were not detected at all. Compared to TEM-type and SHV-type enzymes, CTX-M-type enzymes are more widely disseminated worldwide, and many variants associated with clinically relevant functional heterogeneity have been described. Thus, coexpression of different ESBL types is more common among GNB, which harbors CTX-M-type enzymes [26].
Both the CTX-M-1 group and SHV group were abundantly detected in K. pneumoniae, whereas in E. coli, the SHV group was detected in only 8% of the isolates (Table 8). This finding matches the report by Ogutu et al. that SHV-type is the predominant ESBL enzyme in K. pneumoniae and TEM-type is the most prevalent in E. coli [27].
A single blaNDM-1 carbapenemase gene in a K. pneumoniae isolate and blaNDM-1 plus blaOXA-51 carbapenemase genes in an A. baumannii isolate were detected. The expression of blaNDM-1 in an A. baumannii isolate has previously been reported from the southwestern part of Ethiopia [28], but to our knowledge, no case of blaNDM-1 presence in K. pneumoniae has been reported to date. In general, this finding is not surprising since the blaNDM-1 carbapenemase genes in K. pneumoniae and A. baumannii have been commonly reported from other eastern African countries, such as Kenya, Uganda [29], Egypt [30] and Sudan [31].
In this study, most of the antibiotics initiated by the treating physicians for empiric treatment in the participating patients were ineffective according to the AST. However, these data have to be interpreted with caution since most microbiological culturing was performed after initiation of empiric antibiotic therapy, and evaluation of the clinical success rate of the empirically initiated antibiotic therapies was not part of this investigation. Nevertheless, antibiotic resistance impairing the success of empiric antibiotic therapies seems alarmingly common, and local epidemiological resistance data should be taken into account before the initiation of antibiotic therapy [32]. An adaptation of the local empiric antibiotic therapy strategy could be all the more necessary, as the excessive use of 3GC could be one of the main reasons for the spread of ESBL-producing bacteria [33].
Selectively utilizing antibiotics based on the AST result is not only favorable for optimal treatment success but also plays a major role in combating the spread of MDR bacteria. Adequate microbiological culturing before initiation of empiric antibiotic treatment is necessary to enable AST-based antibiotic therapy. As resources are limited and the supply of laboratory materials is unreliable at the study site, as at many other sites in low-income countries, it may not always be possible to perform comprehensive microbiological testing. In such cases, at least surveillance studies with the subsequent establishment of resistance statistics should be carried out to enable calculated antibiotic therapies adapted to the local resistance status. This might help to reduce the imprudent use of antibiotics [8].
A limitation of this study might be the impaired sensitivity of BCs, since for the first 200 participants, a single locally prepared blood culture bottle was inoculated, and subsequently, only one set of standard commercially available blood culture bottles was used for blood culturing. The limited availability and high cost of commercially available blood culture bottles prevent the regular use of such products on site. Molecular resistance testing was not possible for all gram-negative isolates due to loss upon storage and transport. Our study did not investigate whether infections caused by ESBL- or carbapenemase-producing GNB were associated with reduced success of antibiotic therapies or increased mortality.
Conclusion
In this study, conducted in a tertiary hospital in Ethiopia, we isolated GNB from patients with infectious diseases with high rates of MDR, including 3GC resistance, which is the most commonly used drug class for empiric antibiotic treatment. Based on local AST results, empiric treatment initiated in 72.2% of patients was likely ineffective. As a cause of widespread drug resistance, we found a high prevalence of various ESBL enzymes, with TEM- and CTX-M-1-types predominating. More than half of the gram-negative isolates harbored two or more ESBL genes. In addition, carbapenemases were detected in 7.4% of gram-negative isolates, despite the limited availability and infrequent use of carbapenems in the country. These findings underscore the need for regular microbiological testing of appropriate specimens before initiating empiric antibiotic treatment wherever possible. Therefore, there is an urgent need to strengthen AMS, take appropriate measures to regulate antibiotic use and monitor the emergence of resistant bacteria. In this study, we reported a low positivity rate (5.4%) for BCs, indicating the need for a new diagnostic approach such as plasma cell-free DNA sequencing or the use of MDW- or PCT-guided blood culture to improve the positivity rate.
Availability of data and materials
All relevant data generated or analyzed during this study are included in this article.
Abbreviations
- 3GC:
-
3Rd generation cephalosporin
- AST:
-
Antimicrobial susceptibility testing
- BC:
-
Blood cultures
- CP:
-
Carbapenemase
- CTX-M:
-
Cefotaximase-Munich
- EDTA:
-
Ethylene diamine tetraacetic acid
- ESBL:
-
Extended-spectrum β-lactamase
- EUCAST:
-
European Committee on Antimicrobial Susceptibility Testing
- GNB:
-
Gram-negative bacteria
- IMP:
-
Imipenemase metallo-β-lactamase
- KPC:
-
K. pneumoniae Carbapenemase
- MBL:
-
Metallo-beta-lactamase
- MDR:
-
Multidrug resistance
- MDW:
-
Monocyte distribution width
- NDM:
-
New Delhi metallo-β-lactamase
- OXA:
-
Oxacillinase
- PCR:
-
Polymerase chain reaction
- SHV:
-
Sulfhydryl variable
- SOPs:
-
Standard operating procedures
- TEM:
-
Temoniera
- VIM:
-
Verona imipenemase
References
Misganaw A, Haregu TN, Deribe K, Tessema GA, Deribew A, Melaku YA, Amare AT, Abera SF, Gedefaw M, Dessalegn M, Lakew Y, Bekele T, Mohammed M, Yirsaw BD, Damtew SA, Krohn KJ, Achoki T, Blore J, Assefa Y, Naghavi M. National mortality burden due to communicable, non-communicable, and other diseases in Ethiopia, 1990–2015: findings from the Global Burden of Disease Study 2015. Popul Health Metr. 2017;15:29.
Alosaimy S, Jorgensen SCJ, Lagnf AM, Melvin S, Mynatt RP, Carlson TJ, Garey KW, Allen D, Venugopalan V, Veve M, Athans V, Saw S, Yost CN, Davis SL, Rybak MJ. 2020. Real-world multicenter analysis of clinical outcomes and safety of meropenem-vaborbactam in patients treated for serious gram-negative bacterial infections. Open Forum Infect Dis 7:ofaa051.
CDC. 2019. Antibiotic resistance threats in the United States. Atlanta, GA: US Department of Health and Human Services, CDC; 2019 wwwcdcgov/DrugResistance/Biggest-Threatshtml.
Ruppé É, Woerther PL, Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann Intensive Care. 2015;5:61.
Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72:2145–55.
Martínez-Martínez L, González-López JJ. Carbapenemases in Enterobacteriaceae: types and molecular epidemiology. Enferm Infecc Microbiol Clin. 2014;32(Suppl 4):4–9.
Lester R, Musicha P, van Ginneken N, Dramowski A, Hamer DH, Garner P, Feasey NA. Prevalence and outcome of bloodstream infections due to third-generation cephalosporin-resistant Enterobacteriaceae in sub-Saharan Africa: a systematic review. J Antimicrob Chemother. 2020;75:492–507.
Leekha S, Terrell CL, Edson RS. General principles of antimicrobial therapy. Mayo Clin Proc. 2011;86:156–67.
Fuchs A, Tufa TB, Hörner J, Hurissa Z, Nordmann T, Bosselmann M, Abdissa S, Sorsa A, Orth HM, Jensen BO, MacKenzie C, Pfeffer K, Kaasch AJ, Bode JG, Häussinger D, Feldt T. 2021. Clinical and microbiological characterization of sepsis and evaluation of sepsis scores. PLoS One 16:e0247646.
Moges F, Eshetie S, Abebe W, Mekonnen F, Dagnew M, Endale A, Amare A, Feleke T, Gizachew M, Tiruneh M. 2019. High prevalence of extended-spectrum beta-lactamase-producing Gram-negative pathogens from patients attending Felege Hiwot Comprehensive Specialized Hospital, Bahir Dar, Amhara region. PLoS One 14:e0215177.
Beyene D, Bitew A, Fantew S, Mihret A, Evans M. 2019. Multidrug-resistant profile and prevalence of extended spectrum β-lactamase and carbapenemase production in fermentative Gram-negative bacilli recovered from patients and specimens referred to National Reference Laboratory, Addis Ababa, Ethiopia. PLoS One 14:e0222911.
Strauß LM, Dahms C, Becker K, Kramer A, Kaase M, Mellmann A. Development and evaluation of a novel universal β-lactamase gene subtyping assay for blaSHV, blaTEM and blaCTX-M using clinical and livestock-associated Escherichia coli. J Antimicrob Chemother. 2015;70:710–5.
Wendel AF, Brodner AH, Wydra S, Ressina S, Henrich B, Pfeffer K, Toleman MA, Mackenzie CR. Genetic characterization and emergence of the metallo-β-lactamase GIM-1 in Pseudomonas spp. and Enterobacteriaceae during a long-term outbreak. Antimicrob Agents Chemother. 2013;57:5162–5.
Huang XZ, Cash DM, Chahine MA, Nikolich MP, Craft DW. Development and validation of a multiplex TaqMan real-time PCR for rapid detection of genes encoding four types of class D carbapenemase in Acinetobacter baumannii. J Med Microbiol. 2012;61:1532–7.
Lubwama M, Phipps W, Najjuka CF, Kajumbula H, Ddungu H, Kambugu JB, Bwanga F. Bacteremia in febrile cancer patients in Uganda. BMC Res Notes. 2019;12:464.
Wasihun AG, Wlekidan LN, Gebremariam SA, Dejene TA, Welderufael AL, Haile TD, Muthupandian S. Bacteriological profile and antimicrobial susceptibility patterns of blood culture isolates among febrile patients in Mekelle Hospital. Northern Ethiopia Springerplus. 2015;4:314.
Moges F, Gizachew M, Dagnew M, Amare A, Sharew B, Eshetie S, Abebe W, Million Y, Feleke T, Tiruneh M. Multidrug resistance and extended-spectrum beta-lactamase producing Gram-negative bacteria from three Referral Hospitals of Amhara region. Ethiopia Ann Clin Microbiol Antimicrob. 2021;20:16.
Boyles TH, Davis K, Crede T, Malan J, Mendelson M, Lesosky M. Blood cultures taken from patients attending emergency departments in South Africa are an important antibiotic stewardship tool, which directly influences patient management. BMC Infect Dis. 2015;15:410.
Bassetti M, Russo A, Righi E, Dolso E, Merelli M, Cannarsa N, D’Aurizio F, Sartor A, Curcio F. Comparison between procalcitonin and C-reactive protein to predict blood culture results in ICU patients. Crit Care. 2018;22:252.
Polilli E, Sozio F, Frattari A, Persichitti L, Sensi M, Posata R, Di Gregorio M, Sciacca A, Flacco ME, Manzoli L, Di Iorio G, Parruti G. 2020. Comparison of Monocyte Distribution Width (MDW) and Procalcitonin for early recognition of sepsis. PLoS One 15:e0227300.
Crouser ED, Parrillo JE, Seymour C, Angus DC, Bicking K, Tejidor L, Magari R, Careaga D, Williams J, Closser DR, Samoszuk M, Herren L, Robart E, Chaves F. Improved early detection of sepsis in the ED with a novel monocyte distribution width biomarker. Chest. 2017;152:518–26.
Teklu DS, Negeri AA, Legese MH, Bedada TL, Woldemariam HK, Tullu KD. Extended-spectrum beta-lactamase production and multi-drug resistance among Enterobacteriaceae isolated in Addis Ababa. Ethiopia Antimicrob Resist Infect Control. 2019;8:39.
Abayneh M, Tesfaw G, Abdissa A. Isolation of extended-spectrum β-lactamase- (ESBL-) producing Escherichia coli and Klebsiella pneumoniae from patients with community-onset urinary tract infections in Jimma University Specialized Hospital, Southwest Ethiopia. Can J Infect Dis Med Microbiol. 2018;2018:4846159.
Kettner M, Krćméry V. Development of gentamicin resistance in gram-negative bacteria in Czechoslovakia and correlation with its usage. Drugs Exp Clin Res. 1988;14:511–7.
Wang R, Yang Q, Zhang S, Hong Y, Zhang M, Jiang S. Trends and correlation of antibiotic susceptibility and antibiotic consumption at a large teaching hospital in China (2007–2016): a surveillance study. Ther Clin Risk Manag. 2019;15:1019–27.
D’Andrea MM, Arena F, Pallecchi L, Rossolini GM. CTX-M-type β-lactamases: a successful story of antibiotic resistance. Int J Med Microbiol. 2013;303:305–17.
Ogutu JO, Zhang Q, Huang Y, Yan H, Su L, Gao B, Zhang W, Zhao J, Cai W, Li W, Zhao H, Chen Y, Song W, Chen X, Fu Y, Zhang F. Development of a multiplex PCR system and its application in detection of blaSHV, blaTEM, blaCTX-M-1, blaCTX-M-9 and blaOXA-1 group genes in clinical Klebsiella pneumoniae and Escherichia coli strains. J Antibiot (Tokyo). 2015;68:725–33.
Pritsch M, Zeynudin A, Messerer M, Baumer S, Liegl G, Schubert S, Löscher T, Hoelscher M, Belachew T, Rachow A, Wieser A. First report on bla (NDM-1)-producing Acinetobacter baumannii in three clinical isolates from Ethiopia. BMC Infect Dis. 2017;17:180.
Ssekatawa K, Byarugaba DK, Wampande E, Ejobi F. A systematic review: the current status of carbapenem resistance in East Africa. BMC Res Notes. 2018;11:629.
Khalil MAF, Elgaml A, El-Mowafy M. Emergence of multidrug-resistant New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae in Egypt. Microb Drug Resist. 2017;23:480–7.
Adam MA, Elhag WI. Prevalence of metallo-β-lactamase acquired genes among carbapenems susceptible and resistant Gram-negative clinical isolates using multiplex PCR, Khartoum hospitals. Khartoum Sudan BMC Infect Dis. 2018;18:668.
Montravers P, Dupont H, Gauzit R, Veber B, Bedos JP, Lepape A. Strategies of initiation and streamlining of antibiotic therapy in 41 French intensive care units. Crit Care. 2011;15:R17.
Goyal D, Dean N, Neill S, Jones P, Dascomb K. 2019. Risk Factors for Community-Acquired Extended-Spectrum Beta-Lactamase-Producing Enterobacteriaceae Infections-A Retrospective Study of Symptomatic Urinary Tract Infections. Open Forum Infect Dis 6:ofy357.
Acknowledgements
The authors thank the study and laboratory team of the Hirsch Institute of Tropical Medicine in Asella, Ethiopia, and the staff of the Institute of Medical Microbiology and Hospital Hygiene, Heinrich Heine University, Düsseldorf, Germany for supporting this work.
Funding
The first author of this study, Tafese Beyene Tufa, was supported with stipends by the Bayer Foundation Talents for Africa Scholarship Program and by the Heinz Ansmann Foundation for AIDS research. There was no influence of the funding organization on the analysis or interpretation of the described data.
Author information
Authors and Affiliations
Contributions
TBT: Conception and design, performance of laboratory investigation acquisition, analysis, and interpretation of data, drafting of the manuscript and approval of the manuscript for publication. CM: Interpretation of data, critical revision of the manuscript and approval of the manuscript for publication. HMO: Conception and design, revision of the manuscript and approval of the manuscript for publication. TW: Supervision of the laboratory investigations, interpretation of the results and approval of the manuscript for publication. TN: Conception and design, revision of the manuscript and approval of the manuscript for publication. SA: Performance of laboratory investigations and approval of the manuscript for publication. ZH: Interpretation of data, revision of the manuscript and approval of the manuscript for publication. AS: Conception and design, revision of the manuscript and approval of the manuscript for publication. MB: Conception and design, revision of the manuscript and approval of the manuscript for publication. DH: Interpretation of data, critical revision of the manuscript and approval of the manuscript for publication. KP: Interpretation of data, critical revision of the manuscript and approval of the manuscript for publication. TL: Critical revision of the manuscript and approval of the manuscript for publication. AF: Conception and design, acquisition and interpretation of the data, revision of the manuscript and approval of the manuscript for publication. TF: Conception and design, interpretation and analysis of data, critical revision of the manuscript and approval of the manuscript for publication. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The appropriate ethical review boards of Arsi University, Ethiopia (reference number A/U/H/S/C/120/6443/2017), the Oromia Regional Health Bureau, Ethiopia (reference number BEFO/AHBTFH/1-8/2017), and Düsseldorf University Hospital, Germany (reference number 5729) approved the study. Ethical clearance for sample transportation between Ethiopia and Germany was obtained from the National Ethical Review Board of the Ethiopian Ministry of Science and Technology (reference number 310/204/2017). Before the start of the study procedures, written informed consent to participate in the study was obtained from each study participant or, in the case of minors, from their legal guardians.
Consent for publication
The informed consent form signed by each study participant or, in the case of minors, the legal guardians contained consent for publication of anonymized data.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Tufa, T.B., Mackenzie, C.R., Orth, H.M. et al. Prevalence and characterization of antimicrobial resistance among gram-negative bacteria isolated from febrile hospitalized patients in central Ethiopia. Antimicrob Resist Infect Control 11, 8 (2022). https://doi.org/10.1186/s13756-022-01053-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-022-01053-7