Abstract
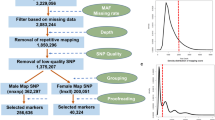
Spinach (Spinacia oleracea L.) is a dioecious plant with male heterogametic sex determination and homomorphic sex chromosomes (XY). The dioecism is utilized for producing commercial hybrid seeds, and hence understanding the molecular-genetic basis of the species’ sex determining locus is an important issue for spinach breeding. In this study, seven dominant DNA markers were shown to completely co-segregate with the male-determining gene in segregating spinach populations comprising > 1500 plants. In addition, these seven dominant DNA markers were completely associated with the male-determining gene in over 100 spinach germplasm accessions and cultivars. These observations suggest that, in spinach, a Y-chromosomal region around the male-determining locus does not (or almost not) recombine with a counterpart region on the X chromosome. Using five of the seven DNA markers, five bacterial artificial chromosome (BAC) clone contigs with a total length of approximately 690 kbp were constructed. Full sequencing of six representative BAC clones (total insert length 504 kbp) from the five contigs and a transcriptome analysis by RNA-seq revealed that the Y-chromosomal region around the male-determining locus contains large amounts of repetitive elements, suggesting that the region might be poor in gene content. Most of the repeats found in this region are novel Ty1-copia-like and its derivative elements that accumulate predominantly in heterochromatic regions. Our findings may provide valuable insight into spinach genome structure and clues for future research into the evolution of the sex determining locus.





Similar content being viewed by others
References
Akamatsu T, Suzuki T, Uchimiya H (1998) Determination of male or female of spinach by using DNA marker. Japanese patent JP 1998052284
Altschul SF, Madden TL, Schäer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bachtrog D (2013) Evolution of Sex Chromosomes. In: Losos J (ed) The Princeton guide to evolution. Princeton University Press, Princeton, pp 387–396
Brandes A, HeslopHarrison J, Kamm A, Kubis S, Doudrick R, Schmidt T (1997) Comparative analysis of the chromosomal and genomic organization of Ty1-copia-like retrotransposons in pteridophytes, gymnosperms and angiosperms. Plant Mol Biol 33:11–21
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden T (2009) BLAST plus: architecture and applications. Bmc Bioinformatics 10:421
Cermak T, Kubat Z, Hobza R, Koblizkova A, Widmer A, Macas J, Vyskot B, Kejnovsky E (2008) Survey of repetitive sequences in Silene latifolia with respect to their distribution on sex chromosomes. Chromosome Res 16:961–976
Charlesworth D (2016) Plant sex chromosomes. Annu Rev Plant Biol 67:397–420
Charlesworth B, Charlesworth D (1978) Model for evolution of dioecy and gynodioecy. Am Nat 112:975–997
Charlesworth B, Sniegowski P, Stephan W (1994) The evolutionary dynamics of repetitive DNA in eukaryotes. Nature 371:215–220
Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, Rupp O, Sörensen TR, Stracke R, Reinhardt R, Goesmann A, Kraft T, Schulz B, Stadler PF, Schmidt T, Gabaldón T, Lehrach H, Weisshaar B, Himmelbauer H (2014) The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature 505:546–549
Ellis JR, Janick J (1960) The chromosomes of Spinacia oleracea. Am J Bot 47:210–214
Fuentes-Bazan S, Mansion G, Borsch T (2012a) Towards a species level tree of the globally diverse genus Chenopodium (Chenopodiaceae). Mol Phylogenet Evol 62:359–374
Fuentes-Bazan S, Uotila P, Borsch T (2012b) A novel phylogeny-based generic classification for Chenopodium sensu lato, and a tribal rearrangement of Chenopodioideae (Chenopodiaceae). Willdenowia 42:5–24
Fujito S, Takahata S, Suzuki R, Hoshino Y, Ohmido N, Onodera Y (2015) Evidence for a common origin of homomorphic and heteromorphic sex chromosomes in distinct Spinacia species. G3 (Bethesda) 5:1663–1673
Geraldes A, Hefer CA, Capron A, Kolosova N, Martinez-Nuñez F, Soolanayakanahally RY, Stanton B, Guy RD, Mansfield SD, Douglas CJ, Cronk QC (2015) Recent Y chromosome divergence despite ancient origin of dioecy in poplars (Populus). Mol Ecol 24:3243–3256
Hammer K (2001) Chenopodiaceae, Spinacia oleracea L. In: Hanelt P, Institut für Pflanzengenetik und Kulturpflanzenforschung Gatersleben Germany (eds) Mansfeld’s encyclopedia of agricultural and horticultural crops (except ornamentals), 1st English edn. Springer, Berlin, pp 249–251
Heslop-Harrison J, Brandes A, Taketa S, Schmidt T, Vershinin A, Alkhimova E, Kamm A, Doudrick R, Schwarzacher T, Katsiotis A, Kubis S, Kumar A, Pearce S, Flavell A, Harrison G (1997) The chromosomal distributions of Ty1-copia group retrotransposable elements in higher plants and their implications for genome evolution. Genetica 100:197–204
Hobza R, Lengerova M, Svoboda J, Kubekova H, Kejnovsky E, Vyskot B (2006) An accumulation of tandem DNA repeats on the Y chromosome in Silene latifolia during early stages of sex chromosome evolution. Chromosoma 115:376–382
Hobza R, Kubat Z, Cegan R, Jesionek W, Vyskot B, Kejnovsky E (2015) Impact of repetitive DNA on sex chromosome evolution in plants. Chromosome Res 23:561–570
Iizuka M, Janick J (1962) Cytogenetic analysis of sex determination in Spinacia oleracea. Genetics 47:1225–1241
Ito M, Ohmido N, Akiyama Y, Fukui K, Koba T (2000) Characterization of spinach chromosomes by condensation patterns and physical mapping of 5S and 45S rDNAs by FISH. J Am Soc Hortic Sci 125:59–62
Janick J (1998) Hybrids in horticultural crops. In: Lamkey KR, Staub JE (eds) Concepts and breeding of heterosis in crop plants. Crop Science Society of America, Madison, pp 45–56
Janick J, Stevenson E (1954) A genetic study of the heterogametic nature of the staminate plant in spinach. Proc Am Soc Hortic Sci 63:444–446
Janick J, Stevenson E (1955) Genetics of the monoecious character in spinach. Genetics 40:429–437
Jukes T, Cantor C (1969) Evolution of protein molecules. In: Munro H (ed) Mammalian protein metabolism. Academic Press, New York, pp 21–132
Kadereit G, Mavrodiev E, Zacharias E, Sukhorukov A (2010) Molecular phylogeny of Atripliceae (Chenopodioideae, Chenopodiaceae): implications for systematics, biogeography, flower and fruit evolution, and the origin of C4 photosynthesis. Am J Bot 97:1664–1687
Kalendar R, Vicient CM, Peleg O, Anamthawat-Jonsson K, Bolshoy A, Schulman AH (2004) Large retrotransposon derivatives: abundant, conserved but nonautonomous retroelements of barley and related genomes. Genetics 166:1437–1450
Kataoka R, Hara M, Kato S, Isobe S, Sato S, Tabata S, Ohmido N (2012) Integration of linkage and chromosome maps of red clover (Trifolium pratense L.). Cytogenet Genome Res 137:60–69
Katayama Y, Shida S (1956) Development of intersexual flowers and their location on the stalk in spinach. Jpn J Breed 6:19–22
Kumar A, Bennetzen J (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Kurtz S, Phillippy A, Delcher A, Smoot M, Shumway M, Antonescu C, Salzberg S (2004) Versatile and open software for comparing large genomes. Genome Biol 5:R12
Liu Z, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu Q, Pearl HM, Kim MS, Charlton JW, Stiles JI, Zee FT, Paterson AH, Ming R (2004) A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature 427:348–352
Na JK, Wang J, Ming R (2014) Accumulation of interspersed and sex-specific repeats in the non-recombining region of papaya sex chromosomes. BMC Genom 15:335
Naeger J, Golenberg E (2016) Mode and tempo of sequence and floral evolution within the Anserineae. Plant Syst Evol 302:385–398
Onodera Y, Yamamoto PM, Kubo T, Tetsuo M (1999) Heterogeneity of the atp6 presequences in normal and different sources of male-sterile cytoplasms of sugar beet. J Plant Physiol 155:656–660
Onodera Y, Yonaha I, Niikura S, Yamazaki S, Mikami T (2008) Monoecy and gynomonoecy in Spinacia oleracea L.: morphological and genetic analyses. Sci Hortic 118:266–269
Onodera Y, Yonaha I, Masumo H, Tanaka A, Niikura S, Yamazaki S, Mikami T (2011) Mapping of the genes for dioecism and monoecism in Spinacia oleracea L.: evidence that both genes are closely linked. Plant Cell Rep 30:965–971
Ossowski S, Schneeberger K, Lucas-Lledó JI, Warthmann N, Clark RM, Shaw RG, Weigel D, Lynch M (2010) The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327:92–94
Picq S, Santoni S, Lacombe T, Latreille M, Weber A, Ardisson M, Ivorra S, Maghradze D, Arroyo-Garcia R, Chatelet P, This P, Terral JF, Bacilieri R (2014) A small XY chromosomal region explains sex determination in wild dioecious V. vinifera and the reversal to hermaphroditism in domesticated grapevines. BMC Plant Biol 14:229
Renner S, Ricklefs R (1995) Dioecy and its correlates in the flowering plants. Am J Bot 82:596–606
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Rogers S, Bendich A (1987) Ribosomal-RNA genes in plants—variability in copy number and in the intergenic spacer. Plant Mol Biol 9:509–520
Sakamoto K, Ohmido N, Fukui K, Kamada H, Satoh S (2000) Site-specific accumulation of a LINE-like retrotransposon in a sex chromosome of the dioecious plant Cannabis sativa. Plant Mol Biol 44:723–732
Sassa H (2007) A technique to isolate DNA from woody and herbaceous plants by using a silica-based plasmid extraction column. Anal Biochem 363:166–167
Stanke M, Diekhans M, Baertsch R, Haussler D (2008) Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24:637–644
Takahata S, Yago T, Iwabuchi K, Hirakawa H, Suzuki Y, Onodera Y (2016) Comparison of spinach sex chromosomes with sugar beet autosomes reveals extensive synteny and low recombination at the male-determining locus. J Hered 107:679–685
Trapnell C, Pachter L, Salzberg S (2009) TopHat: discovering splice junctions with RNA-SEq. Bioinformatics 25:1105–1111
VanBuren R, Zeng F, Chen C, Zhang J, Wai CM, Han J, Aryal R, Gschwend AR, Wang J, Na JK, Huang L, Zhang L, Miao W, Gou J, Arro J, Guyot R, Moore RC, Wang ML, Zee F, Charlesworth D, Moore PH, Yu Q, Ming R (2015) Origin and domestication of papaya Yh chromosome. Genome Res 25:524–533
Vyskot B, Hobza R (2015) The genomics of plant sex chromosomes. Plant Sci 236:126–135
Wang J, Na JK, Yu Q, Gschwend AR, Han J, Zeng F, Aryal R, VanBuren R, Murray JE, Zhang W, Navajas-Pérez R, Feltus FA, Lemke C, Tong EJ, Chen C, Wai CM, Singh R, Wang ML, Min XJ, Alam M, Charlesworth D, Moore PH, Jiang J, Paterson AH, Ming R (2012) Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci USA 109:13710–13715
Westergaard M (1958) The mechanism of sex determination in dioecious flowering plants. Adv Genet Incorp Mol Genet Med 9:217–281
Wicker T, Keller B (2007) Genome-wide comparative analysis of copia retrotransposons in Triticeae, rice, and Arabidopsis reveals conserved ancient evolutionary lineages and distinct dynamics of individual copia families. Genome Res 17:1072–1081
Xu Z, Wang H (2007) LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res 35:W265–W268
Xu C, Jiao C, Sun H, Cai X, Wang X, Ge C, Zheng Y, Liu W, Sun X, Xu Y, Deng J, Zhang Z, Huang S, Dai S, Mou B, Wang Q, Fei Z (2017) Draft genome of spinach and transcriptome diversity of 120 Spinacia accessions. Nat Commun 8:15275
Yamamoto K, Oda Y, Haseda A, Fujito S, Mikami T, Onodera Y (2014) Molecular evidence that the genes for dioecism and monoecism in Spinacia oleracea L. are located at different loci in a chromosomal region. Heredity 112:317–324
Acknowledgements
The authors thank Tohoku Seed Co. Ltd. (Utsunomiya, Tochigi, Japan) very much for providing the spinach breeding lines used in this study. We appreciate the technical assistance provided by Mrs. H. Yokomoto. The work presented here was supported by the Interuniversity Bio-Backup Project (IBBP) as an application code (Hokkaido 0001). We used the DNA Sequencing Facility of Research Faculty of Agriculture, Hokkaido University.
Funding
This work was supported by the Japan Society for the Promotion of Science (Grants-in-Aid for Scientific Research (KAKENHI) Grant Numbers 26292001 and 16H06279), Ministry of Education, Culture, Sports, Science and Technology (KAKENHI Grant Number 221S0002), and the Takeda Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data availability
The BAC clone sequences (AP017636–AP017641) and the RNA-seq reads (DRA006107) generated during and/or analyzed during the current study are available in the DNA Data Bank of Japan (DDBJ). The primer sequences of the SP_0018 marker developed by Tohoku Seed Co., Ltd. is not publicly available because it remains a trade secret.
Additional information
Communicated by S. Hohmann.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kudoh, T., Takahashi, M., Osabe, T. et al. Molecular insights into the non-recombining nature of the spinach male-determining region. Mol Genet Genomics 293, 557–568 (2018). https://doi.org/10.1007/s00438-017-1405-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-017-1405-2