Abstract
Background
Long non-coding RNAs (LncRNAs) have recently been considered promising biomarkers for oncogenesis due to their epigenetic regulatory effects. HOTAIR is one of the oncogenic LncRNAs that was previously studied in different non-hematological malignancies. The current study set out to detect the expression level of HOTAIR LncRNA in AML patients concerning their clinical characteristics, laboratory data, FLT3-ITD, and NPM1 mutations, as well as treatment outcome. This study included quantitative detection of HOTAIR gene expression in 47 cases of AML using quantitative reverse transcription polymerase chain reaction, as well as NPM1 and FLT3-ITD genotyping.
Results
The HOTAIR expression was significantly higher in AML patients 6.87 (0.001) than in normal controls 1.66 (0.004–6.82) (p 0.007). The HOTAIR expression level was affected by chemotherapy, and it was correlated to hemoglobin level (p 0.001), age, total leukocytic count (p 0.022), and NPM1 mutation (p 0.017). HOTAIR gene expression level showed a correlation to relapse-free survival in the study group (p 0.04).
Conclusion
HOTAIR is overexpressed in patients with acute myeloid leukemia (AML). HOTAIR pre-treatment and post-chemotherapy gene expression levels can predict chemosensitivity and relapse.
Similar content being viewed by others
Background
The availability of whole-genome sequencing enabled discovering the genomic landscape and the non-coding genes, representing ~ 98% of the human genome [1, 2]. Long non-coding RNAs (LncRNAs) are non-coding RNA transcripts that exceed 200 base pairs (bp) in length. LncRNAs are located and transcribed from different genomic loci and are designated as intergenic and intronic LncRNAs from intergenic regions and introns, respectively [3].
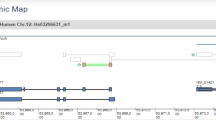
HOX transcript antisense intergenic RNA (HOTAIR) is a 2158-bp LncRNA transcribed from the antisense strand of the homeobox C gene on chromosome 12 [4]. HOTAIR acts in trans on the HOX D locus of chromosome 2 through chromatin-remodeling enzymes [5]. It is a novel prognostic biomarker, especially for the potential metastatic prediction [6]. The HOTAIR overexpression has been shown in patients with colorectal cancer and hepatocellular carcinoma [7]. It has been reported that the degree of HOTAIR expression has an impact on the survival rate of breast cancer patients [8]. It has been demonstrated that inhibiting HOTAIR promotes cancer cell apoptosis [9].
The Fms-like tyrosine kinase three-gene (FLT3) encodes a tyrosine kinase expressed on hematopoietic progenitor cells. FLT3 is the most common mutated gene in AML [10]. The most common FLT3 mutation in AML is an internal tandem duplication (FLT3-ITD) in the juxtamembrane domain of the FLT3 gene, resulting in uncontrolled hematopoietic cell proliferation mediated by FLT3/STAT5 [11].
The Nucleophosmin 1 gene (NPM1) mutations have been described as one of the most frequent genetic lesions in AML, occurring in about 40% of normal karyotype AML [12]. The NPM1 gene encodes a nucleo-cytoplasmic shuttling protein that regulates the ARF-p53 tumor-suppressor pathway [13]. NPM1 mutations result in an abnormal cytoplasmic accumulation of the NPM1 protein [14]. NPM1 mutations have been associated with distinct LncRNA signatures in AML [15].
The present study aims to determine the HOTAIR expression level in AML patients using quantitative real-time reverse transcription PCR and correlate the expression level with NPM1 and FLT-ITD mutations, as well as laboratory data and treatment outcome.
Methods
This research aims to examine the HOTAIR gene expression in AML and the association between HOTAIR expression and clinicopathological data from patients, including the commonly tested mutations in our lab (e.g., NPM1 and FLT3-ITD).
The study included 47 patients with AML from Medical Oncology Department. The post-chemotherapy level of HOTAIR gene expression was compared to pre-treatment levels and treatment outcome.
The controls were age- and sex-matched volunteers referred to the Clinical Pathology Department for bone marrow analysis before splenectomy for hypersplenism. Informed consent was obtained from all subjects for being included in the study.
The following are the inclusion criteria:
-
All cases of AML above 18 years old (adults)
-
AML with recurrent cytogenetic abnormalities
-
AML NOS
-
Both males and females
The following are the exclusion criteria:
-
Pediatric age group
-
Therapy-related AML
-
AML with myelodysplasia-related changes
-
History of myeloproliferative neoplasms (e.g., chronic myeloid leukemia or primary myelofibrosis)
The study included the following:
-
Detection of NPM1 and FLT3-ITD mutations by high-resolution melting (HRM) analysis
-
Quantitative detection of HOTAIR gene expression
-
Follow-up of cases after treatment (standard AML chemotherapy 7 + 3 protocol)
-
HOTAIR gene expression level detection in cases with adequate post-chemotherapy bone marrow samples (adequate RNA extract)
NPM1 and FLT3-ITD mutation screening by high-resolution melting analysis
Genomic DNA was extracted from bone marrow samples using the QIAamp® DNA Blood Mini Kit (Cat No:51104). Twenty-five nanogram of the DNA was used in the PCR reaction, which was performed using MeltDoctor™ HRM Master Mix (ABI Product Number/4415440). The primers that had been used in the genotyping of NPM1 and FLT3-ITD were as follows: NPM1 forward 5′-TGGTTCCTTAACCACATTTCTTT-3′ and reverse 5′-GGACAACACATTCTTGGC-3′ [16], while for FLT3-ITD: forward 5′-TGCAGAACTG CCTATTCCTAACTGA-3′ and reverse 5′-TTCCATAAGCTGTTGCGTTCATCA-3′ [17]. The reaction plate was run using the Applied Biosystems 7500 Fast Real-Time PCR under the following conditions: holding 95 °C for 10 min, 50 cycles of denaturation at 95 60 °C for 15 s, and annealing/extension at 60 °C for 1 min. The melting curve conditions were as follows: (i) denaturation at 95 °C for 30 s, (ii) annealing at 60 °C for 1 min, (iii) denaturation at 95 °C for 30 s, and (iv) annealing at 60 °C for 15 s [16, 17].
HOTAIR gene expression
Total RNA was extracted from bone marrow mononuclear cells using the QlAamp RNA Blood Mini Kit (Cat No. 52304), followed by cDNA synthesis using the RevertAid First Strand cDNA Synthesis Kit (Cat No: K1622). HOTAIR gene expression was performed using the RT qPCR technique, with b-actin serving as a housekeeping gene. SYBR Green qPCR Master Mix (Cat No: K0221) was used. The primers that had been used in the gene expression experiment were as follows: HOTAIR, forward 5′-CAGTGGGGAACTCTGACTCG-3′ and reverse 5′-GTGCCTGGTGCTCTCTTACC-3′; β-actin, forward 5′-CACCATTGGCAATGAGCGGTTCC-3′ and reverse 5′-GTAGTTTCGTGG ATGCCACAGG-3′ [18]. The reaction conditions were as follows: (i) holding at 95 °C for 10 min, (ii) 10 cycles of denaturation at 95 °C for 15 s, (iii) annealing/extension at 95-60 °C (touch-down) for 1 min, (iv) another 40 cycles of denaturation at 95 °C for 30 s, (v) annealing at 56 °C for 1 min, and (vi) extension at 72 °C for 30 s. The expression was calculated using the 2−ΔΔCT formula, where ΔCT = Ct HOTAIR − Ctβ-actin and ΔΔCT = ΔCT sample − ΔCT control (reference sample) [19].
Statistical analysis
Statistical analysis was carried out using SPSS v25. The Kruskal-Wallis test was used to compare cases and controls, and Pearson correlation was used to associate the clinicopathological features with HOTAIR gene expression. P values < 0.05 are considered significant. Survival rates (overall survival and disease-free survival) were calculated by Kaplan Mayer (log-rank) analysis.
Results
The study involved 47 participants, 26 males and 21 females. Their age ranged from 18 to 83 years, with a median of 36 years. According to the FAB classification, 29 cases were M2 and M4. The summary of hematological data is shown in Table 1.
Results of NPM1 and FLT3-ITD mutation screening by HRM
HRM analysis was done using the ABI7500 fast HRM software version 2.02. Previous studies validated the technique of HRM for detecting NPM1 and FLT3-ITD [16, 17].
Figures 1 and 2 illustrate the melting curves and difference plots between controls, wild genotype, and mutant genotypes. Wild genotype in both genes was found in 22 cases. In 5 cases, mutations in both genes were found. Five cases had NPM1 mutant and FLT3-ITD wild genotypes, 14 cases had NPM1 wild and FLT3-ITD mutant genotypes, and 1 case was not examined for NPM1 or FLT3-ITD mutations (failed DNA extraction).
HOTAIR detection of gene expression in AML cases and controls
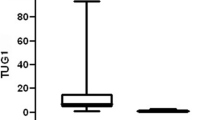
There was a substantial difference in the median HOTAIR gene expression levels between cases and controls (Kruskal-Wallis, p 0.007). HOTAIR expression in cases varied from 0.001 to 2702 with a median expression of 6.87, while it ranged from 0.004 to 6.82 with a median expression of 1.66 in controls, as illustrated in Table 2.
The correlation of HOTAIR expression level to age, gender, and different hematological data was illustrated in Table 3. High HOTAIR levels were associated with higher hemoglobin (Hb) levels, lower total leucocytic count (TLC), and lower age groups. Other clinicopathological factors, including age, gender, platelet count (PLT), and complete remission (CR), were not significantly correlated. Regarding NPM1 and FLT3 mutation status, all the NPM1 mutant/FLT3-ITD wild cases expressed low levels of HOTAIR (p = 0.017). Thirty-seven cases started to receive standard induction therapy; 18/37 cases achieved CR, while the remaining cases were deemed unfit to start induction.
HOTAIR expression in AML patients prior and post-chemotherapy (16/37) (see Table 4)
Cases with a high expression demonstrated a substantial reduction after CR (p = 0.028). However, cases with a low HOTAIR expression group showed a non-significant increase in HOTAIR expression following CR. Patients who failed to achieve CR, whether they had low or high HOTAIR expression, exhibited a non-significant decrease in HOTAIR gene expression after induction.
Combined effect of HOTAIR expression and NPM1/FLT3-ITD mutation analysis on CR achievement and poor outcome (see Table 5)
The NPM1 mutant/FLT3-ITD mutant group (100%) achieved the highest CR rate among the low expression group, while the lowest rate was found in the NPM1-wild/FLT3-ITD mutant group (16%). Among the high expression group, CR was maximally achieved in the NPM1-wild/FLT3-ITD mutant (63%). Poor outcome was defined in the study by death, relapse, and failed induction. The level of HOTAIR expression did not affect the poor outcome percentages except in the NPM1-wild/FLT3-ITD mutant genotype, where the high expression was associated with diminished poor outcome percentages.
Overall survival
According to Table 6, Kaplan-Meier survival analysis revealed that patients in the lower TLC group and those who achieved CR had a better overall survival (OS) (p = 0.05, p = 0.004, respectively).
Relapse-free survival
The 18 patients who achieved CR after induction therapy were monitored for 12 months to assess relapse-free survival (RFS). RFS factors such as age, gender, Hb, PLT, blasts percent, HOTAIR expression, and NPM1/FLT3 gene mutation status and data are summarized in Table 7. Seven patients relapsed during the follow-up period, with a median RFS of 9 months.
HOTAIR expression levels had a significant outcome on RFS; patients with a high HOTAIR expression showed a higher RFS rate than those with low HOTAIR expression levels (p 0.04). The median RFS was 11 months in the high HOTAIR expression group, compared to 7 months in the low expression group. Regarding the NPM1 and FLT3-ITD mutation screening results, the low HOTAIR expression group in NPM1-WT/FLT3-WT cases (n = 4) had a higher RFS than the high expression group (n = 2). In all cases with FLT3-ITD, the high HOTAIR expression group had a higher RFS than the low expression group.
Discussion
Mutations and misregulation of long non-coding RNAs have been shown to play pivotal roles in oncogenesis. Alterations in LncRNA expression or mutations promote tumorigenesis and metastasis. LncRNAs carry considerable promise as cancer novel biomarkers and therapeutic targets [20].
Histones epigenetic modifications, such as acetylation and methylation, have been proved to be frequently deregulated in AML [21].
The effect of methylation is site-specific, for example, histone 3 lysine 4 (H3K4), activates gene expression. On the contrary, methylation of histone 3 lysine 9 (H3K9) or lysine 27 (H3K27) suppresses the transcription [22].
The PRC2 complex contains the three core protein subunits: enhancer of zeste homolog 2 (EZH2), embryonic ectoderm development, and SUZ12. EZH2 contains a SET domain that catalyzes trimethylation of histone H3 at lysine 27. HOTAIR binding to PRC2 is required for H3K27 trimethylation [23].
Through whole-genome sequencing, NPM1 mutations and FLT3-ITD were found to be associated with distinct LncRNA signatures [24].
HOTAIR was reported to act as a bridge between protein complexes, supporting chromatin silencing, including the PRC2 [5].
HOTAIR binds to and sequesters miR-193a competitively, modulates the expression of c-KIT in AML cells, promotes cell growth, and inhibits apoptosis [25].
Patients with NPM1 mutations proved to have a better outcome with increased complete remission and improved overall survival [26]. However, AML with FLT3 ITD carries a poor prognosis owing to its higher risk of relapse [27].
In this study, the expression of LncRNA and HOTAIR genes in 47 AML cases was estimated using the RT qPCR technique compared to controls. HOTAIR gene expression was higher in AML cases than in controls. These findings match those of a previous study, reporting that HOTAIR was overexpressed in AML patients [28].
According to HOTAIR median expression, the patients were classified into high and low expression groups. The high expression group showed a significant decrease after CR achievement (p = 0.024). These results mirror those of previous studies, which found that the HOTAIR expression was reduced after the achievement of CR [18]. However, a non-significant increase in the HOTAIR expression levels was noticed in the low expression group after CR achievement.
HOTAIR expression was affected by NPM1 and FLT3 mutations. All NPM1 mutant/FLT3-ITD wild cases were from the low HOTAIR expression group (p = 0.017), and 40% of the double NPM1/FLT3-ITD mutant cases were from the low expression group.
Since all NPM1 mutations increased cytoplasmic concentrations of NPM1 protein, NPM1 modulates stress response and growth suppression by binding to and stabilizing p53 and inhibiting MDM2 [29].
Zhai et al. had demonstrated that the p53 level was negatively correlated with HOTAIR in non-small cell lung carcinoma, suggesting a regulatory role for p53 in HOTAIR transcription. They discovered two p53 binding sites in the HOTAIR promoter region [30]. Similarly, the strong negative correlation between HOTAIR expression level and NPM1 mutation can be explained.
The HOTAIR expression was found to be positively correlated with hemoglobin level, which contradicts a previous study [18].
However, Shih and Kung explained that HOTAIR expression was upregulated by hypoxia, which is a common feature of all malignancies, including AML. This upregulation occurs through the hypoxia-responsive element in the HOTAIR promoter region, which is activated directly by hypoxia-induced factor-1 α (HIF1α) [31]. HIF was found to upregulate δ-aminolevulinate synthase, which catalyzes the first and rate-limiting reaction in the heme biosynthesis [32]. TLC was found to be inversely associated with HOTAIR expression (p = 0.022). Previous studies on TLC and HOTAIR expression levels were controversial. Some researchers rated the TLC as high and low according to the highest normal count (10 × 109/L) and found a positive correlation between HOTAIR expression and TLC [18].
However, Zhang et al. concluded that there is no correlation between the HOTAIR expression and TLC of AML patients. They classified the cases into high and low TLC groups by the count 100 × 109/L [33].
In this study, no significant correlation was found between patients’ age, gender, platelet count, CR achievement, and HOTAIR expression.
CR achievement correlation with combined HOTAIR expression and mutation status revealed that among the low expression group, CR was maximally achieved in the NPM1 mutant/FLT3-ITD mutant genotype (100%) and minimally in the NPM1 wild/FLT3-ITD mutant group. In contrast, among the high expression group, CR was maximally achieved in the NPM1-wild /FLT3-ITD mutant.
Just 20% of the NPM1-mutant/FLT-wild party achieved CR, which is probably explained by other risk factors. Two patients had died before ending their induction, one patient was not fit to start therapy, and one patient missed his follow-up.
Patients in the NPM1 wild/FLT3-ITD mutant group experienced an unexplained rise in CR rates, despite having two weak prognostic factors: the FLT3-ITD mutation and elevated levels of HOTAIR oncogenic LncRNA expression.
Göllner et al. found that when FLT3-ITD mutation-positive leukemic cell lines after they have been cultured in media containing the kinase inhibitor for an extended time, they developed resistance to the kinase inhibitor and standard chemotherapeutics. Western blot identified complete loss of H3K27me3 in resistant cells. They also established that the mechanism of action is not restricted to the FLT3-ITD mutation [34].
The increased CR achievement rates with high HOTAIR expression in the FLT3-ITD-Mutant group were due to the interaction between HOTAIR LncRNA and its PRC2 binding function, which allows trimethylation of H3K27 and thus promotes chemo-sensitivity even in the presence of FLT3 ITD.
In this study, the OS of patients showed a high positive association with CR achievement (p = 0.004) and a low TLC (p = 0.05). These findings corroborate a previous study [18].
The HOTAIR high expression group had a greater RFS (p = 0.024), surprisingly against previous literature [2]. This finding was corroborated with the post-chemotherapy HOTAIR expression analysis, in which we found that the low expression group after achieving CR had higher HOTAIR levels than at diagnosis, and 3/4 of patients relapsed during follow-up, implying that post-treatment HOTAIR expression levels may serve as an early predictor of the relapse susceptibility.
Poor outcome was found in all combinations of NPM1and FLT3-ITD results regardless of the HOTAIR expression level except patients with FLT3-ITD mutant/NPM1-wild populations, where the poor outcome ameliorated from 84% in patients with low HOTAIR expression levels to 62% in patients with high HOTAIR expression levels. However, this improvement was not statistically important. It can be attributed to our finding of increased chemo-sensitivity in the group with high HOTAIR expression in the presence of FLT3-ITD mutation.
Conclusion
In conclusion, HOTAIR expression levels were significantly higher in AML patients than in normal controls. According to the median of HOTAIR expression, patients were divided into high and low groups. Expression in the high group was markedly reduced after induction therapy and CR achievement. HOTAIR expression level correlated strongly with the Hb level at diagnosis, but not with the presence of the favorable mutation group (NPM1 mutation-FLT3 IDT wild-type), age, and TLC. Follow-up of patients revealed that our patients’ overall survival was affected by TLC and CR achievement. A higher relapse rate was found in the low HOTAIR expression group.
Availability of data and materials
All data will be available upon request after acceptance.
Informed consent was obtained from cases and controls.
Abbreviations
- AML:
-
Acute myeloid leukemia
- ARF:
-
ADP-ribosylation factor
- CR:
-
Complete remission
- CT:
-
Threshold cycle
- EED:
-
Embryonic ectoderm development
- EZH2:
-
Enhancer of zeste homolog 2
- FAB:
-
French-American-British
- FLT3-ITD:
-
Fms-like tyrosine kinase 3 gene internal tandem duplication
- H3K4:
-
Histone 3 lysine 4
- H3K9:
-
Histone 3 lysine 9
- H3K27:
-
Histone 3 lysine 9
- Hb:
-
Hemoglobin
- HIF1α:
-
Hypoxia-induced factor-1 α
- HOTAIR:
-
HOX transcript antisense intergenic RNA
- HRM:
-
High-resolution melting analysis
- LncRNAs:
-
Long non-coding RNAs
- MDM2:
-
Mouse double minute 2 homolog
- OS:
-
Overall survival
- PRC:
-
Polycomb repressive complex 2
- P53:
-
Tumor protein P53
- RFS:
-
Relapse-free survival
- SUZ12:
-
Suppressor of zeste 12 homolog
- TLC:
-
Total leucocytic count
References
Zhang X, Meyerson M (2020) Illuminating the noncoding genome in cancer. Nature Cancer 1(9):864–872. https://doi.org/10.1038/s43018-020-00114-3
Diamantopoulos MA, Tsiakanikas P, Scorilas A (2018) Non-coding RNAs: the riddle of the transcriptome and their perspectives in cancer. Ann Transl Med 6(12):241. https://doi.org/10.21037/atm.2018.06.10
Grixti JM, Ayers D (2020) Long noncoding RNAs and their link to cancer. Non-Coding RNA Res 5(2):77–82. https://doi.org/10.1016/j.ncrna.2020.04.003
Jin K-T, Lu Z-B, Lv J-Q, Zhang J-G (2020) The role of long non-coding RNAs in mediating chemoresistance by modulating autophagy in cancer. RNA Biol 17(12):1727–1740. https://doi.org/10.1080/15476286.2020.1737787
Yuan C, Ning Y, Pan Y (2020) Emerging roles of HOTAIR in human cancer. J Cell Biochem 121(5–6):3235–3247. https://doi.org/10.1002/jcb.29591
Toy HI, Okmen D, Kontou PI, Georgakilas AG, Pavlopoulou A (2019) HOTAIR as a prognostic predictor for diverse human cancers: a meta- and bioinformatics analysis. Cancers 11(6):778. https://doi.org/10.3390/cancers11060778
Kim JO, Jun HH, Kim EJ, Lee JY, Park HS, Ryu CS, Kim S, Oh D, Kim JW, Kim NK (2020) Genetic variants of HOTAIR associated with colorectal cancer susceptibility and mortality. Front Oncol 10:72. https://doi.org/10.3389/fonc.2020.00072
Li C, Cui J, Zou L, Zhu L, Wei W (2020) Bioinformatics analysis of the expression of HOXC13 and its role in the prognosis of breast cancer. Oncol Lett 19(1):899–907. https://doi.org/10.3892/ol.2019.11140
Liu Y, Chen X, Chen X, Liu J, Gu H, Fan R, Ge H (2020) Long non-coding RNA HOTAIR knockdown enhances radiosensitivity through regulating microRNA-93/ATG12 axis in colorectal cancer. Cell Death Dis 11(3):175. https://doi.org/10.1038/s41419-020-2268-8
Kiyoi H, Kawashima N, Ishikawa Y (2020) FLT3 mutations in acute myeloid leukemia: therapeutic paradigm beyond inhibitor development. Cancer Sci 111(2):312–322. https://doi.org/10.1111/cas.14274
Bazarbachi A, Bug G, Baron F, Brissot E, Ciceri F, Dalle IA, Döhner H, Esteve J, Floisand Y, Giebel S, Gilleece M, Gorin NC, Jabbour E, Aljurf M, Kantarjian H, Kharfan-Dabaja M, Labopin M, Lanza F, Malard F, Peric Z, Prebet T, Ravandi F, Ruggeri A, Sanz J, Schmid C, Shouval R, Spyridonidis A, Versluis J, Vey N, Savani BN, Nagler A, Mohty M (2020) Clinical practice recommendation on hematopoietic stem cell transplantation for acute myeloid leukemia patients with FLT3-internal tandem duplication: a position statement from the Acute Leukemia Working Party of the European Society for Blood and Marrow Transplantation. Haematologica 105(6):1507–1516. https://doi.org/10.3324/haematol.2019.243410
Bacher U, Porret N, Joncourt R, Sanz J, Aliu N, Wiedemann G, Jeker B, Banz Y, Pabst T (2018) Pitfalls in the molecular follow up of NPM1 mutant acute myeloid leukemia. Haematologica 103(10):e486–e488. https://doi.org/10.3324/haematol.2018.192104
Mitrea DM, Kriwacki RW (2018) On the relationship status for Arf and NPM1 – it’s complicated. FEBS J 285(5):828–831. https://doi.org/10.1111/febs.14407
Kunchala P, Kuravi S, Jensen R, McGuirk J, Balusu R (2018) When the good go bad: mutant NPM1 in acute myeloid leukemia. Blood Rev 32(3):167–183. https://doi.org/10.1016/j.blre.2017.11.001
Papaioannou D, Petri A, Dovey OM, Terreri S, Wang E, Collins FA, Woodward LA, Walker AE, Nicolet D, Pepe F, Kumchala P, Bill M, Walker CJ, Karunasiri M, Mrózek K, Gardner ML, Camilotto V, Zitzer N, Cooper JL, Cai X, Rong-Mullins X, Kohlschmidt J, Archer KJ, Freitas MA, Zheng Y, Lee RJ, Aifantis I, Vassiliou G, Singh G, Kauppinen S, Bloomfield CD, Dorrance AM, Garzon R (2019) The long non-coding RNA HOXB-AS3 regulates ribosomal RNA transcription in NPM1-mutated acute myeloid leukemia. Nat Commun 10(1):5351. https://doi.org/10.1038/s41467-019-13259-2
Huet S, Jallades L, Charlot C, Chabane K, Nicolini FE, Michallet et al (2013) New quantitative method to identify NPM1 mutations in acute myeloid leukaemia. Leukemia Res Treat:1–5. https://doi.org/10.1155/2013/756703
Tan AYC, Westerman DA, Carney DA, Seymour JF, Juneja S, Dobrovic A (2008) Detection of NPM1 exon 12 mutations and FLT3 - internal tandem duplications by high resolution melting analysis in normal karyotype acute myeloid leukemia. J Hematol Oncol 1:10. https://doi.org/10.1186/1756-8722-1-10
Wu S, Zheng C, Chen S, Cai X, Shi Y, Lin B, Chen Y (2015) Overexpression of long non-coding RNA HOTAIR predicts a poor prognosis in patients with acute myeloid leukemia. Oncol Lett 10(4):2410–2414. https://doi.org/10.3892/ol.2015.3552
Rao X, Huang X, Zhou Z, Lin X (2013) An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinform Biomath 3(3):71–85 https://pubmed.ncbi.nlm.nih.gov/25558171
Gutschner T, Richtig G, Haemmerle M, Pichler M (2018) From biomarkers to therapeutic targets—the promises and perils of long non-coding RNAs in cancer. Cancer Metastasis Rev 37(1):83–105. https://doi.org/10.1007/s10555-017-9718-5
Dhall A, Zee BM, Yan F, Blanco MA (2019) Intersection of epigenetic and metabolic regulation of histone modifications in acute myeloid leukemia. Front Oncol 9:432 https://www.frontiersin.org/article/10.3389/fonc.2019.00432, 9
Li J, Ahn JH, Wang GG (2019) Understanding histone H3 lysine 36 methylation and its deregulation in disease. Cell Mol Life Sci 76(15):2899–2916. https://doi.org/10.1007/s00018-019-03144-y
Martin CJ, Moorehead RA (2020) Polycomb repressor complex 2 function in breast cancer (review). Int J Oncol 57(5):1085–1094. https://doi.org/10.3892/ijo.2020.5122
Papaioannou D, Nicolet D, Volinia S, Mrózek K , Yan P. Bundschuh, R, et al., (2017). Prognostic and biologic significance of long non-coding RNA profiling in younger adults with cytogenetically normal acute myeloid leukemia. Haematologica, 102(8), 1391–1400 doi: https://doi.org/10.3324/haematol.2017.166215
Tang Q, Hann SS (2018) HOTAIR: an oncogenic long non-coding RNA in human cancer. Cell Physiol Biochem 47(3):893–913. https://doi.org/10.1159/000490131
Box J, Paquet N, Adams M, Boucher D, Bolderson E, O’Byrne K et al (2016) Nucleophosmin: from structure and function to disease development. BMC Mol Biol 17(1):19. https://doi.org/10.1186/s12867-016-0073-9
Gaballa S, Saliba R, Oran B, Brammer JE, Chen J, Rondon G, Alousi AM, Kebriaei P, Marin D, Popat UR, Andersson BS, Shpall EJ, Jabbour E, Daver N, Andreeff M, Ravandi F, Cortes J, Patel K, Champlin RE, Ciurea SO (2017) Relapse risk and survival in patients with FLT3 mutated acute myeloid leukemia undergoing stem cell transplantation. Am J Hematol. 92(4):331–337. https://doi.org/10.1002/ajh.24632
Wang S-L, Huang Y, Su R, Yu Y-Y (2019) Silencing long non-coding RNA HOTAIR exerts anti-oncogenic effect on human acute myeloid leukemia via demethylation of HOXA5 by inhibiting Dnmt3b. Cancer Cell Int 19(1):114. https://doi.org/10.1186/s12935-019-0808-z
Vu TT, Stölzel F, Wang KW, Röllig C, Tursky ML, Molloy TJ, Ma DD (2020) miR-10a as a therapeutic target and predictive biomarker for MDM2 inhibition in acute myeloid leukemia. Leukemia. https://doi.org/10.1038/s41375-020-01095-z
Zhai N, Xia Y, Yin R, Liu J, Gao F (2016) A negative regulation loop of long noncoding RNA HOTAIR and p53 in non-small-cell lung cancer. Onco Targets Ther. 9:5713–5720. https://doi.org/10.2147/OTT.S110219
Botti G, Scognamiglio G, Aquino G, Liguori G, Cantile M (2019) LncRNA HOTAIR in tumor microenvironment: what role? Int J Mol Sci 20(9). https://doi.org/10.3390/ijms20092279
Stojanovski BM, Hunter GA, Na I, Uversky VN, Jiang RHY, Ferreira GC (2019) 5-Aminolevulinate synthase catalysis: the catcher in heme biosynthesis. Mol Genet Metab 128(3):178–189. https://doi.org/10.1016/j.ymgme.2019.06.003
Zhang Y-Y, Huang S-H, Zhou H-R, Chen C, Tian L-H, Shen J-Z (2016) Role of HOTAIR in the diagnosis and prognosis of acute leukemia. Oncol Rep 36(6):3113–3122. https://doi.org/10.3892/or.2016.5147
Göllner S, Oellerich T, Agrawal-Singh S, Schenk T, Klein H, Rohde C et al (2017) Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med 23(1):69–78. https://doi.org/10.1038/nm.4247
Acknowledgements
Part of this work was published as a poster presentation and in the abstract book in Abstracts from the 2019 Lymphoma and Myeloma Congress October 23–26, 2019, New York, NY, November 2019
Funding
The study was funded by Cairo University (as post-graduate research).
Author information
Authors and Affiliations
Contributions
All authors contributed equally. MS put the aim of the research and the methodology. HZ was the clinical oncologist, who was concerned by the treatment and clinical follow-up. NB and NS had edited and revised the manuscript. FA is the corresponding author who collected the data, performed the PCR experiments, and performed the statistical analysis. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by both clinical pathology and clinical oncology departments as well as post-graduate committee as part of the MD thesis fulfillment; the reference number is I-351015. Written informed consent was obtained from all subjects included in the study.
Consent for publication
No identifiable data were used in the study. However, written informed consent for participation was obtained from all participants.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Salah, M., Zawam, H., Fouad, N.B. et al. Study of HOTAIR LncRNA in AML patients in context to FLT3-ITD and NPM1 mutations status. Egypt J Med Hum Genet 22, 61 (2021). https://doi.org/10.1186/s43042-021-00180-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s43042-021-00180-x