Abstract
Background
Increasing the presence of beneficial soil microorganisms is a promising sustainable alternative to support conventional and organic fertilization and may help to improve crop health and productivity. If the application of single bioeffectors has shown satisfactory results, further improvements may arise by combining multiple beneficial soil microorganisms with natural bioactive molecules.
Methods
In the present work, we investigated in a pot experiment under greenhouse conditions whether inoculation of two phosphate-solubilizing bacteria, Pseudomonas spp. (B2) and Bacillus amyloliquefaciens (B3), alone or in combination with a humic acids (HA) extracted from green compost and/or a commercial inoculum (M) of arbuscular mycorrhizal fungi (AMF), may affect maize growth and soil microbial community. Phospholipid fatty acid (PLFA) and denaturing gradient gel electrophoresis (DGGE) fingerprinting analysis were performed to detect changes in the microbial community composition.
Results
Plant growth, N and P uptake, and mycorrhizal root colonization were found to be larger in all inoculated treatments than in the uninoculated control. The greatest P uptake was found when B. amyloliquefaciens was applied in combination with both HA and arbuscular mycorrhizal fungi (B3HAM), and when Pseudomonas was combined with HA (B2HA). The PLFA-based community profile revealed that inoculation changed the microbial community composition. Gram+/Gram− bacteria, AMF/saprotrophic fungi and bacteria/fungi ratios increased in all inoculated treatments. The greatest values for the AMF PLFA marker (C16:1ω5) and AMF/saprotrophic fungi ratio were found for the B3HAM treatment. Permutation test based on DGGE data confirmed a similar trend, with most significant variations in both bacterial and fungal community structures induced by inoculation of B2 or B3 in combination with HA and M, especially in B3HAM.
Conclusions
The two community-based datasets indicated changes in the soil microbiome of maize induced by inoculation of B2 or B3 alone or when combined with humic acids and mycorrhizal inoculum, leading to positive effects on plant growth and improved nutrient uptake. Our study implies that appropriate and innovative agricultural management, enhancing the potential contribution of beneficial soil microorganisms as AMF, may result in an improved nutrient use efficiency in plants.

Similar content being viewed by others
Background
The intensive use of mineral fertilizers and chemical pesticides has been driving the agricultural productivity of the past century. Future challenges for agriculture require innovative cropping technologies for a more efficient management of the limited natural resources to preserve soil fertility and minimize the adverse environmental impact of current agricultural production [1]. In this regard, beneficial plant–microbe interactions can be manipulated to improve crop production in agriculture. Focusing on microbiome engineering is an emerging biotechnological strategy of a sustainable intensification in agriculture [2].
In soil–plant systems, prebiotics are products as organic amendments, usually manure, compost, biochar or humic substances, promoting the growth of probiotics, soil microorganisms already present within the soil–plant system or favoring the establishment of inoculated microorganisms. In the same field, probiotics are specifically considered as beneficial microorganisms, which improve plant fitness and nutrient availability. Use of soil amendments as prebiotics and inoculation with specific probiotics microorganisms represent promising tools to develop and establish a beneficial plant microbiome [2].
Organic fertilizers, such as compost, have the advantage of helping to recycle parts of nutrients, that are already available in the agroecosystem, enriching soil with organic matter that contains nutrients in organic forms less readily accessible to plant uptake and less susceptible to leaching [3, 4]. Thus, their application may increase the nutrient use efficiency, that is one of the final goals of a sustainable intensification of agriculture.
In this context, bioeffectors (BEs), including various plant growth-promoting microorganisms (PGPM) and/or bioactive natural compounds support important functions in sustainable agriculture as supplements to conventional fertilizers; they appear as important boosters of soil fertility enhancing the efficiency of nutrients recycling in the soil [5]. Among microbial BEs, Bacillus and Pseudomonas species are common and abundant bacterial populations in the rhizosphere of various crops [64], suggesting a high competence to colonize plant surfaces and tissues. Strains belonging to these genera provide numerous beneficial traits, such as root growth promotion, solubilization of sparingly soluble nutrients and stimulation of root colonization by mycorrhizal fungi [6, 7]. This last trait plays a key functional role by mediating the transfer of carbon from roots to soil as a source of energy for microbial life and contribute to plant uptake of mineral nutrients and water [8]. However, none of the known PGPM per se has the potential to fulfill the requested requirements of providing a viable alternative to mineral fertilizers [9]. In fact, it has been recently recognized that the interactive and synergistic effects expected from PGPM cannot be achieved with a single application [10, 11].
Mycorrhizal fungi are described as crucial in soil systems since they establish a mutualistic association with the majority of plants [12]. AMF diversity in agricultural soils has been strongly reduced by conventional agricultural practices [13], including overuse of fertilizers, agrochemicals, tillage, long fallow periods. AMF diversity and abundance in agricultural soils could be improved by changing management practices which promote indigenous AMF population, or by reintroducing them through inoculation [14]. The multiple services provided by AMF are the result of the synergistic activities of the bacterial communities living in the mycorrhizosphere, including N fixation, P solubilization, and the production of phytohormones, siderophores, and antibacterial metabolites [15]. Cooperation among beneficial microorganisms has been confirmed for different species of phosphate-solubilising bacteria (e.g., Bacillus spp., Pseudomonas spp.) in diverse plant species [15,16,17,18,19,20].
Besides synergistic interactions among microbial bioeffectors, a combination with natural bioactive compounds holding plant growth-promoting effects can also provide additional benefits for plant growth. Humic substances (HS) have attracted rising attention by farmers and scientific community owing to their capacity to regulate many ecological and environmental processes and can be used as soil conditioning agents. In fact, HS sustain plant growth and microbial life controlling soil C and N cycling, growth of plants and microorganisms, plant root initiation and architecture, and stabilization of soil structure [21,22,23,24,25,26,27,28]. Several studies reported the synergistic effects of the integrated use of HS plus biofertilizers on plant yield and nutrient uptake [29]. The anatomical and physiological changes in roots induced by HS may favor the fitness of the mutualistic interaction by increasing rhizosphere population and chemotaxis, bacteria attachment and survival on plant surface as well as endophytic colonization [30]. Moreover, several authors have reported that the combination of microbial inoculants with HS gives more reproducible results for plant growth and production [31,32,33]. They may represent a key component of a new generation of biofertilizers acting as carriers of microbial inoculants in soil [34]. The application of different microbial inoculant species together in consortia or plus HS has been proposed as a strategy to increase the fitness of the inoculants when confronted with the complex and highly competitive native soil community [34, 35]. In addition, the application of humus-rich organic fertilizers, such as compost, has shown the ability of PGPM to improve nutrient uptake and plant growth [36,37,38,39,40,41,42].
This study was performed within the activities of the EU project BIOFECTOR that aimed to test different microbial strains, commercially available, able to promote plant growth, P solubilization and arbuscular mycorrhiza helper functions. The specific objectives of our research were to test whether the combined application of humic acids (HA) and microbial bioeffectors Pseudomonas spp. DSMZ 13134 (B2), Bacillus amyloliquefaciens FZB42 (B3), alone or in combination with a mix of arbuscular mycorrhizal fungi (Rhizoglomus irregulare and Funneliformis mosseae) can promote synergy and more efficacy, on maize plant growth and nutrient uptake. Further, to have a deeper understanding of the effects of microbial bioeffectors alone or in combination with HA on plant performances, we examined whether different combination may affect microbial community composition and mycorrhizal root colonization in the presence of a native microbial community in a non-sterilized soil. Our hypothesis was that the combined application of microbial bioeffectors and HA would show greater benefits for the crop than individual application of microbial strains, and concomitantly affect the composition of soil microbial communities.
Materials and methods
Soil and compost material
The surface layer (0–15 cm) of a soil classified as Vertic Xerofluvent was collected at the Experimental Farm of the University of Naples Federico II located in Castel-Volturno (CE), Italy. The sampled soil was air-dried and sieved to < 5 mm before being analyzed for the content of organic C by the Walkley–Black method [43], total N by the Kjeldahl digestion method [44], and the available soil P by the sodium bicarbonate Olsen method [45]. The soil showed a clay loam texture (44.6%, 28% and 27.4% sand, silt and clay, respectively), an alkaline pH of 8.6 (1:2.5 soil:water) and contained 1.11 g kg−1 total N, 10.5 g kg−1 organic carbon, 11 mg kg−1 of NaHCO3-extractable P.
The green compost as a source of humic acids (HA) was produced at the on-farm composting facility of the Experimental Farm of the University of Naples Federico II. The composting process was based on a static pile with air insufflation system, formed by a rotative pump connected to a frame of perforated rubber tubes. The tubes were placed on a bed of dry corn residues (4 × 8 m). The composting pile was made up by a mixture (base matrix) of cow and buffalo manure (70% w/w) and artichoke residues as structuring woody material (30% w/w); the mixed material was uniformly spread by a power shovel to cover the insufflation system and forming the final pile height of approximately 1.5 m. The composting lasted 100 days, with a periodic monitoring of external and internal temperature level (5 min interval) and oxygen percentage (60 min interval). During the first 50 days, the minimum percentage of oxygen was set at 10%, then subsequently at 5%. Compost was oven-dried at 40 °C until constant weight and sieved at 500 μm. Dry compost samples were analyzed by a Fisons EA 1108 (Fisons Instruments, Milano, Italy) elemental analyzer and revealed an elemental content of 27.9, 2.13, and 4.06% of C, N, and H, respectively, and a C/N ratio of 13.1.
Bioeffectors
The bioeffectors consisted of three microbial inocula and one HA. The commercial microbial products were: Proradix® WP (Pseudomonas spp. DSMZ 13,134 containing 5.0 × 1010 colony-forming units g−1) (B2) produced by Sourcon Padena Tubingen, Germany; RhizoVital® 42 fl. (Bacillus amyloliquefaciens FZB42, also referred as Bacillus velezensis FZB42, containing 2.5 × 1010 spores g−1) (B3) produced by ABiTEP GmbH, Berlin, Germany; Aegis Sym Microgranule based on a mixture of two arbuscular mycorrhizal fungi strains of Rhizophagus irregularis BEG72 and Funneliformis mosseae BEG234, containing 25 spores g−1 of each strain, produced by, Agrotecnologias Naturales Atens, S.L. Tarragona, Spain. Stock suspensions of each bacterial product were prepared under sterile conditions using demineralized water with 2.5 mM CaSO4 and sprayed on seeds surface at sowing both at the rate of 2 × 106 cfu per g of substrate. Aegis Sym was applied directly as microgranule at the rate of 2.5 g pot−1 at sowing.
HA were extracted from green compost based on artichoke residues as follows: 100 g of air-dried compost (2 mm sieved) was suspended in 1000 mL of a 0.1 mol L−1 NaOH and 0.1 mol L−1 Na4P2O7 solution and mechanically shaken for 24 h. The suspension was then centrifuged at 7500 g for 20 min and filtered through glass wool. The extraction was repeated twice after 1 h of shaking. The filtered supernatants were acidified to pH 1.5 with 6 mol L−1 HCl to precipitate the humic acids. After 24 h, samples were centrifuged at 2500 g for 20 min and the HA collected and dialysed (1 kD cutoff Spectrapore tubes) against deionized water until the electrical conductivity became lower than 0.5 dS m−1. Stock solution was prepared from freeze-dried powder dissolved in deionized water and diluted to the application rate (0.012 g Kg−1). In an earlier study, we performed the chemical characterization of HA isolated from artichoke compost and tested their bioactivity of towards young maize plants [46].
Pot experiment
The pot experiment was conducted under open greenhouse conditions (25–33 °C, daily temperature range during maize growth,) for 8 weeks starting from May to July. Maize (Zea mays, cv Aphoteos, Limagrain S.p.a.) plants were grown in pots filled with 5 kg of a soil, prepared by mixing the Vertic Xerofluvent clay-loam soil with quartz sand at a 2:1 ratio. A manure on-farm compost (manure: straw, 70:30% w/w) was used as fertilizer (140N-50P-190 K mg kg−1), by mixing it homogenously into the soil, and allowing the mixture to equilibrate for 3 weeks at room temperature before the experiment.
The pot trial was designed as follows:
B0 | Control (no inoculation) |
B0M | Control + mycorrhizal inoculum |
B2 | Pseudomonas spp. |
B2M | Pseudomonas spp. + mycorrhizal inoculum |
B2HA | Pseudomonas spp. + HA |
B2HAM | Pseudomonas spp. + HA + mycorrhizal inoculum |
B3 | Bacillus amyloliquefaciens |
B3M | Bacillus amyloliquefaciens + mycorrhizal inoculum |
B3HA | Bacillus amyloliquefaciens + HA |
B3HAM | Bacillus amyloliquefaciens + HA + mycorrhizal inoculum |
Each treatment was replicated 5 times for a total of 50 pots. Three maize seeds were sown 3 cm below the soil surface in each pot and 15 mL solutions of each BE were applied using an automatic pipette with sterile tips into each planting hole. Mycorrhizal inoculum was placed under the seeds. Holes were closed and the top surface of the soil were covered with 150 g (~ 0.5-cm -hick layer) of quartz sand (0.6–1.2 mm) to avoid the formation of surface crusts after watering. After emergence (5 days after sowing), only one maize seedling per pot was then left to grow and additional 15 mL of BE (bacteria and HA) solution was applied to the top layer. During crop growth, soil water content was maintained at 60% of field capacity by periodically adding water to compensate evapotranspiration losses. The plant performances were monitored weekly by recording plants height, while symptoms of nitrogen deficiencies were corrected, if needed, by supplying 0.5 g pot−1 N as Ca(NO3)2 Plants were harvested 8 weeks after sowing. Plant height and leaf numbers were measured, as well as shoot and root separated, and fresh and dried biomass, after drying in a forced-air oven at 70 °C for 2 days. The soil strictly adhering to roots was considered as rhizosphere and separated by brushing after a gentle shake. The rest of soil was considered bulk soil. Both soils were sieved at 2 mm; rhizosphere soil was stored at − 20 °C for PLFA and molecular fingerprinting analysis, while bulk soil was air-dried and stored for physical–chemical analyses. Total P in maize shoots was obtained by digesting the dried plant samples with diluted HNO3 and HCl (1:3 v/v) [47] and then colorimetrically determined by the molybdenum blue assay method [45]. Total nitrogen was determined by the Kjeldahl digestion method [44].
Mycorrhizal colonization
Root mycorrhizal colonization was estimated by a modified method of Vierheilig [48]. Maize roots were washed with tap water, cleared in 10% KOH for 10 min at 90 °C in a water bath, rinsed in water, and then soaked in 2 M HCl for few minutes. After soaking, the roots were stained in a 5% ink-vinegar solution (5% v/v ink in a 5% acetic acid solution) for 5 min at 90 °C in a water bath. The roots were then rinsed with acidified tap water. Mycorrhizal colonization was estimated by the gridline intersect method [49].
Fatty acid analyses
Selected fatty acids pertaining to the soil phospholipid (PLFA), and used as biomarkers for specific soil microbial communities, were extracted using the modified Bligh and Dyer technique [50], as previously described [51]. Total soil lipids were extracted from 4 g of soil by a chloroform/methanol (MeOH)/citrate buffer (1:2:0.8 v/v). Separation of lipid classes was conducted in silica gel columns by sequentially eluting neutral, glyco-, and phospholipid fractions with chloroform, acetone, and methanol, respectively. The eluted phospholipid fractions were dried under a N2 flow at 37 °C and stored at − 20 °C. Fatty acid methyl esters were formed by a mild alkaline methanolysis. Thirty microliters of methyl nonadecanoate fatty acid (19:0; Sigma-Aldrich) were added as internal standard, and the methylated samples were dried under N2 flow. These samples were redissolved in 200 μl of hexane. Then, for each sample, 2.5 μl was injected in splitless mode by the injector held at 250 °C into a Perkin-Elmer Autosystem XL (GC) equipped with a PE Turbomass-Gold quadrupole mass spectrometer. Chromatographic separation was achieved through a 60 m Supelco Capillary column (SLB-5 ms) using helium as carrier gas (1 mL min−1). The initial oven temperature, 100 °C, was held for 5 min, raised to 210 °C at a rate of 2 °C min−1, then raised from 210 to 250 °C at a rate of 5 °C min−1, and held for 12 min. Mass spectra were obtained in EI mode (70 eV), scanning in the range of m/z 50–600, with a cycle time of 1 s. The abundance of individual PLFA was derived from the relative area under each chromatographic peak, as compared to that of the internal standard (19:0) and related to the calibration curve of the 19:0 standard fatty acid dissolved in hexane.
Total PLFA was calculated as the sum of all phospholipid fatty acids and considered to represent microbial biomass. Each PLFA content was expressed as ng of PLFA per g of dry soil. Fatty acids were named according to the ω-designation as it follows: total number of carbons followed by a colon; the number of double bonds; the symbol ω; and the position of the first double bond from the methyl end of the molecule. Cis- and trans-configurations are indicated by c and t, respectively; iso and anteiso forms of methyl-branched fatty acids are indicated by i- and a-, respectively. 10Me indicates a methyl group placed on the tenth C atom from the carboxyl end of the molecule; cy refers to cyclopropane fatty acids. The C18:2ω6c and C18:1ω9c PLFA were used as biomarkers for fungal biomass [52], while C16:1ω5 PLFA was the biomarker for arbuscular mycorrhizal fungi [53], and it can be used for evaluating the amount of extraradical mycelia of AMF [54]. The aC15:0, iC15:0, iC16:0, iC17:0, and aC17:0 PLFA were chosen to represent Gram+ bacteria, while the C16:1ω7c, C18:1ω7c, C18:1ω5c, cyC17:0, and cyC19:0 PLFA were related to Gram- bacteria [55]. The PLFAs 10Me16:0 10MeC17:0 and 10Me18:0 were used to indicate soil actinomycetes [56]. Structure of soil microbial community was also evaluated using the PLFA ratios relative to bacterial/fungal, Gram+/Gram− bacteria, and AMF/saprotrophic fungi.
Total DNA (TC-DNA) extraction
Total DNA (TC-DNA) was extracted using four out of five replicates subjected to TC-DNA extraction (each 0.5 g of wet weight) after a harsh lysis step, using FastPrep FP24 bead-beating Instrument. The procedure was repeated twice, (30 s, 5.5 m s−1) (MP Biomedicals), by means of the FastDNA spin kit for soil (MP Biomedicals). Purification of extracted TC-DNA was performed using GeneClean spin kit (MP Biomedicals) according to the manufacturer protocol. DNA yield was checked by agarose gel electrophoresis (0.8% w/v agarose; 100 V, 40 min) using ethidium bromide staining and estimated using the 1-Kb Plus DNA ladder. Purified DNA was stored at − 20 °C.
Amplification of bacterial 16S rRNA gene fragments
The purified TC-DNA was diluted 1:10 with deionized water before polymerase chain reaction (PCR) amplification. F984GC/R1378 universal bacterial primer pair was used to amplify TC-DNA as described by Heuer [57]. All primers used in this study are reported in Table 1. The PCR master buffer consisted of: 1 μL of template DNA (1–5 ng), 5 μL GoTaq Flexi Buffer, 3.5 μL MgCl2 (25 mM), 2 μL acetamide (50%), 2.5 μL deoxynucleotide triphosphates (dNTPs, 2 mM), 0.5 μL primers (10 μM), 0.125 μL GoTaq polymerase (5 u/μL), and 9.625 μL deionized water. The PCR program included an initial denaturation step at 94 °C for 5 min, followed by 35 thermal cycles of 94 °C for 1 min, 53 °C for 1 min, and 72 °C for 2 min and a final extension at 72 °C for 10 min.
Amplification of fungal ITS fragments
The fungal community was studied based on the fungal internal transcribed spacer (ITS) fragment and was amplified using a nested PCR approach, as described by Weinert [58] with ITS1F/ITS4 primer sets [59]. The reaction mixture for the first PCR was composed of: 1 μL of template DNA, 2.5 μL TrueStart Buffer, 2.5 μL dNTPs (2 mM), 3.75 μL MgCl2 (25 mM), 0.5 μL primers (10 μM), 0.5 μL DMSO, 0.125 μL TrueStart Taq polymerase (5u/μL), 13.6 μL of deionized water. Thermal cycle started with denaturation at 95 °C for 5 min, followed by 30 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 1 min and a final extension at 72 °C for 10 min. Samples were used as templates for the second PCR. The buffer for the second PCR was the same as the one used for the first PCR, except for the use of 0.2 μL TrueStart Taq polymerase and different primer set (ITS1F-GC/ITS2). The PCR conditions were the same as those described for the first PCR except for the number of cycles, which were reduced to 25. Final amplified samples were checked by means of electrophoresis agarose gel prior to further DGGE analysis.
Analysis of 16S rRNA gene and ITS fragments by denaturing gradient gel electrophoresis (DGGE)
DGGE analysis was performed using a PhorU2 apparatus (Ingeny, Goes, The Netherlands), according to Weinert [58]. Two different double gradient gels were used to separate gene fragments: a gel composed of 46.5–65% denaturants (100% denaturant was defined as 7 M urea and 40% formamide) and acrylamide (6.2–9%) was used for both community and group-specific 16S rRNA, meanwhile the gradient consisted of 23–58% denaturant and 8% acrylamide for fungal gene fragments. The gels were run in Tris-acetate-EDTA buffer and voltage was kept constant at 140 V for 17 h at 58 °C for bacterial community and 100 V for 18 h at 60 °C for fungal community. After electrophoresis the gels were silver stained as described by Heuer [60], air-dried and scanned for acquiring digital pictures. DGGE profiles were processed using GelCompar II software ver. 6.5 (Applied Maths) as described by Rademaker [61] and modified by Smalla [62].
Cluster analysis of DGGE fingerprint patterns was carried out using pairwise Pearson correlation coefficients calculated by means of unweighted pair group method using arithmetic averages (UPGMA). Pearson similarity matrices were used to test significant differences between treatments by the application of permutation test with PERMTEST software (10,000 simulations), calculating the d-value (dissimilarity value) from the average overall correlation coefficients within the groups minus the average overall correlation coefficients between groups from treatments compared, in accordance with Kropf [63].
Statistical analyses
The significant differences between mean values were determined by a one-way analysis of variance, while application of Tukey’s test to differentiate among results was given at the P < 0.05 probability level using the Statgraphics Centurion software version XV. Data were checked for normality and homogeneity of variance and transformed where necessary. Multivariate principal component analysis was used to analyze PLFA biomarkers by means of XLSTAT software (Version 2014.4.08, Addinsoft, Inc., Brooklyn, NY, USA). Pearson’s correlation coefficients were used to test the relationships between the PLFA microbial markers in soil, N and P content in plant tissues, and mycorrhizal root colonization.
Results
Plant growth, nutrient content and percentage of root mycorrhizal inoculation
Maize plants inoculated with bioeffectors, alone or in combination, significantly increased total dry biomass compared to the control (B0) (Fig. 1a). There were small differences among the inoculated treatments but significant, except for B3 that showed the smallest value with an increased biomass of only 13%, as compared to control, whereas in other treatments the increase was from 30% (B2HAM) to 44% (B3HA, B2HA, B2M, and B2) (Fig. 1a). Increase in N and P uptake was also observed in inoculated plants compared to non-inoculated control plants (Fig. 1b, c). The trend was quite similar to that of dry plant biomass, with small differences among the inoculated treatments. In fact, the shoot N content was about 39% larger than control in B3HA and B3HAM treatments, followed by B2M and B3M (with 34% increase), whereas the smallest value (only 9.8% increase) was observed in B3 (Fig. 1b). Phosphorus content was greatest in plants from the B3HAM treatment with a 59% increase over control, followed by that for B2HA, B3M, B3HA, and B2HAM (with about a 40% increase) (Fig. 1c).
Total dried shoot maize biomass (a), shoot N content (b), shoot P content (c), and root colonization by AMF (d) as affected by different treatments. Bars indicate standard deviation of means (n = 5). Different letters above the bars indicate significantly different means according to Tukey’s test (P < 0.05)
Roots were well colonized by AMF in all treatments, being the percentage of root colonization from 65 to 82% (Fig. 1d). In general, inoculated plants had a greater mycorrhizal colonization than control (Fig. 1d). The B3HAM, B3M, B3HA, B2M, and B2HA treatments showed the largest percentage values of root colonization with slight differences among the treatments.
Fatty acid analysis
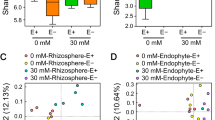
The observed PLFA profiles differed significantly among treatments (Table 2). However, the addition of different bioeffectors, alone or in combination, did not result in significant differences in total microbial biomass based on PLFA analyses, except for the B2 treatment that reached the greatest value (355.9 ng g−1) (Table 2) and the B2HAM with the smallest values (280 ng g−1). A similar trend was observed for the total bacterial biomass. Nevertheless, several significant shifts were observed in some microbial groups. In particular, the saprotrophic fungal biomass was greater in B0 than for the inoculated treatments, except for B2. Conversely, the abundance of the specific AMF C16:1ω5 PLFA marker was greater than B0 control in all inoculated treatments, but particularly in all B3 treatments with a significant increase ranging from 46 to 236% and with the greatest value (61.2 ng g−1) reached for the B3HAM treatment (Table 2). The PLFA results related to Gram+ bacteria showed greater values in all B2 treatments (about 27% in B2) than in control and B3M, B3HA and B3HAM treatments (Table 2). Conversely, the values found for Gram− bacteria did not change significantly among treatments. However, the Gram+ to Gram− bacterial ratios were greater than control for all inoculated treatments, indicating an increase of the relative abundance of Gram+ bacteria (Fig. 2a). A similar trend was observed for bacterial to fungal PLFA ratio (Fig. 2b) and for the ratio of AMF to saprotrophic fungal biomass, as shown by the C16:1ω5/(C18:2ω6,9 + C18:1ω9) ratio that reached the largest value in the B3HAM treatment (Fig. 3c).
PLFA ratios in control and inoculated treatments soils: Gram-positive (Gram+)/Gram-negative (Gram−) (a); bacteria/Fungi (b); AMF/saprotrophic fungi (c). Bars indicate standard deviation of means (n = 5). Different letters above the bars indicate significantly different means according to Tukey’s test (P < 0.05)
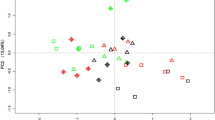
PCA biplot showing the loadings for the different microbial groups and ratios [Gram+/Gram− (G+/G−), total PLFA bacterial/total saprotrophic fungi (bact/fungi) and AMF/saprotrophic fungi (AMF/fungi)] as assessed by PLFA analysis. a Combination of PC1 with PC2; b combination of PC1 with PC3. The combination of the shapes and colors indicate different treatments. Triangle: single inoculation; circle: combination with AMF inoculum (M); diamond: combination with humic acids (HA); square: combination with humic acids plus AMF inoculum (HAM)
A correlation between microbial groups and treatments was achieved by applying a multivariate principal component analysis (PCA) (Fig. 3a, b). The PCA of the PLFA profiles of microbial groups clearly separated the treatments with Pseudomonas or Bacillus, alone or in combination with either mycorrhizal inoculum or HA. The first two PC1 and PC2 components (Fig. 3a) accounted for 74.33% of the total variance, while PC1 and PC3 components (Fig. 3b) did for 70.52%. PC1 (51% of variance) showed negative loadings for AM fungi, but positive loadings for Gram− and saprotrophic fungi, thus suggesting an opposite correlation between these microbial groups. Moreover, the placement of B3M, B3HA and B3HAM along the PC1 suggested an overall positive correlation with AM fungi, Gram+/Gram− and bacterial/fungal ratios. The negative loadings along the PC2 (23% of variance) were associated to B0 and correlated to saprotrophic fungi, whereas the negative loadings along the PC3 (19% of variance), were associated to B3M, B0M and B2HAM and correlated to Gram+/Gram− ratio. It is noteworthy that B3HAM was completely separated from all other treatments and strongly correlated to AMF and AMF/saprotrophic fungi ratio (Fig. 3b).
The correlation among nutrients content, root mycorrhizal colonization and the fatty acid microbial markers provided additional information (Table 3). In fact, P content was positively correlated with AMF marker, AMF/saprotrophic fungi ratio and mycorrhizal root colonization. An opposite correlation was instead found with saprotrophic fungi and actinomycetes. Nitrogen content showed also a similar positive correlation with AMF marker, whereas the negative correlation with saprotrophic fungi was not significant.
PCR-DGGE fingerprint analysis
The analysis of DGGE fingerprints by UPGMA revealed that the bacterial community composition of the different treatments shared approximately 40% similarity (Fig. 4; Additional file 1: Figure S1). All replicates clustered according to their own treatment delineating consistent differences among treatments. In general, the strongest effects were observed when single bacterial strains were combined with the mycorrhizal inoculum and/or with HA. According to pairwise comparisons (Table 4), B3HAM was the treatment with a major shift in the bacterial community composition (d = 82.95, P < 0.05), as well as B3HA (d = 67.51 P < 0.05), although quite similar differences were observed for all combination treatments inoculated with B2. Mycorrhizal inoculum alone (B0M) did not modify the bacterial community composition, although the d-values calculated on pairwise comparisons (Table 4) suggested a strong shift when it was combined with B2 (d = 46.2 P < 0.05), markedly greater than when B2 was applied alone (d = 9.34 P < 0.05).
Fungal communities gel showed a remarkable diversity of bands but also suggested a high level of internal variability (Additional file 1: Figure S2). A cluster profile of inoculant B3 group treatments was clearly visible, showing how this group strongly affected fungal structural composition in soil (Fig. 5). The B3 inoculation effect was confirmed by the permutation test that showed large dissimilarity values when treatments inoculated with Bacillus were compared to control (Table 4). However, also in this case, it is noteworthy that both inoculated treatments showed major differences upon combination of bioeffectors, particularly for B2HA, B3HA and B3HAM (Table 4) while, as shown also for bacterial community composition, the mycorrhizal inoculum alone did not affect fungal community composition (Table 4).
Discussion
Plant growth, nutrient uptake and mycorrhizal colonization
Plant growth and nutrient uptake
We screened two different plant growth-promoting bacterial inoculants either alone or combined with the application of HA isolated from a green artichoke compost and a mycorrhizal inoculum and evaluated maize growth using composted manure as fertilizer, under non-sterilized soil conditions. Our results suggested an overall capacity of both bacterial BEs, inoculated either alone or in combination with HA and mycorrhizal inoculum, to positively affect plant growth and nutrient uptake. These outcomes indicated that the introduced microorganisms, in not sterile soil conditions, exploited the applied compost and the nutrients contained therein more than the indigenous microbial communities. Hence, the present study confirms earlier findings which showed that the inoculation of Pseudomonas spp. and B. amyloliquefaciens alone enabled an improved plant growth and nutrient uptake [37,38,39, 64]. Positive effects were revealed also by the addition of the mycorrhizal inoculum alone, as widely reported in the literature [10, 11, 65].
In the case of bacterial inoculation alone, the Pseudomonas spp. strain showed a better performance in terms of maize plant growth, and N and P uptake than Bacillus amyloliquefaciens. This is in line with previous reports of Thonar et al. [38] and Li et al. [37] on alkaline soil under a compost fertilization. In the latter work, it was shown that the two bacterial strains increased plant P uptake by promoting a fast mineralization and exploitation of compost, thus favoring a rapid microbial growth, and enhanced cycling and fluxes of available P over time, as compared to treatments without inoculation. In fact, most of the species belonging to the genera Bacillus and Pseudomonas are able to access organic P forms, indicating their crucial role in the P cycle of soils [66]. As recently reported by Eltbany et al. [64], inoculation with Pseudomonas spp. and B. amyloliquefaciens FZB42 increased alkaline phosphomonoesterase activity, suggesting an accelerated excretion of P cycling enzymes.
However, when we applied Pseudomonas in combination with HA or mycorrhizal inoculum, there were no significant differences in terms of improved biomass or nutrient uptake in respect to B2 alone, except for P content in plant shoots in the B2HA treatment. On the contrary, although B. amyloliquefaciens alone resulted less beneficial effect to maize plants than Pseudomonas, its combined B3M, B3HA and B3HAM treatments showed an improved plant growth, N and P uptake compared to B3 alone and the best absolute performance in terms of P uptake in B3HAM treatment. Several studies indicated that the combination of Bacillus or Pseudomonas strains with AMF inocula has beneficial effects on plant development [18, 19, 67]. Adesemoye [68] found that mixture of B. amyloliquefaciens and AMF at 70% of fertilization rate produced the same yield as the full fertilization rate without inoculants. As well, a synergistic interaction between Funneliformis mosseae and Pseudomonas fluorescens provided benefits to maize plants in field conditions [11]. Such bacteria can form biofilm by producing exopolysaccharides, which allow them to efficiently colonize roots and mycorrhizal hyphae [69]. However, different bacterial strains may have positive, negative or neutral effect when combined with specific AMF strains on plant performance [10, 18, 70,71,72]. The synergistic effects between the two microbial groups could depend on whether the bacteria are able to solubilize soil P and enhance plant P uptake mediated by AMF. A positive effect of P solubilizing bacteria on extraradical AMF hyphal growth can help them to access new area of mycorrhizosphere and increase access by AMF hyphae to new sources of solubilized P [18]. Interestingly, Battini et al. [19] reported that several Bacillus strains may facilitate root growth and P uptake by favoring the development of AMF extraradical mycelium. They underlined that the larger shoot P content in bacterial inoculated plants was less related to phosphate solubilization than to bacterial induction of greater root length and soil hyphal abundance, which were promoted by the production of indole acetic acid (IAA), as recently reported in several Bacillus species [42, 67, 72, 73]. Thus, in our study, the synergistic or no effect observed could be the result of different capacity of bacterial strains to release IAA, stimulate root and hyphal growth, increase the soil exploitation and/or interact with other microbial actors in the mycorrhizosphere.
Humic acids were recognized to exert a powerful synergic action with Bacillus spp. and Pseudomonas strains increasing plant nutrients uptake, chlorophyll content and growth [74,75,76,77]. In our study, we found a synergistic effect, when HA were combined with each bacterial strain, especially in improving P uptake, compared to B2 or B3 alone. Soil microorganisms are able to mediate complex relationship between HA and P. Bioactive molecules as HA may induce changes in root morphology and architecture, promoting an increase of both area and length of root systems and root hairs [28], favoring the extension of bacteria association with root surface and thus their persistency and activity [34]. These structural interactions between bacteria and plants, in the presence of HA, may explain the reinforced positive effects on P uptake for the combination of microbial inoculants and HA observed in our study. Moreover, it has been reported a coordinated action between HS and microbial inoculants, involving the biosynthetic pathway of IAA that may affect P availability [30]. HS may increase root exudation and the release of precursor compounds of IAA as L-Tryptophan that acts as IAA precursor both in plants and microbes, thus enhancing IAA content in the rhizosphere with a consequent localized acidification and the release of solubilized phosphate in soil solution [78].
Mycorrhizal colonization
It has been already shown that AMF colonization of plant roots is favored by mycorrhiza helper bacteria, such as various Pseudomonas sp. and Pseudomonas fluorescens strains and Bacillus strains [6, 11, 18,19,20, 72, 79]. Our results showed that all inoculated treatments increased root mycorrhizal colonization, even if not always statistically significant. However, inoculation with AMF inoculum slightly increases mycorrhizal root colonization over the control treatment that contained a native AMF community. Interestingly, the two bacterial strains differentially affected mycorrhizal root colonization also in presence of a native AMF community. This indicates not only that the commercial AM inoculum used in the present study enhanced root colonization, but also that the two bacterial strains stimulated a positive response of the indigenous arbuscular mycorrhizal fungi community. Accordingly, Ordonez [18] reported that different strains of Pseudomonas strongly affected colonization of potato roots by AMF, and this effect was more pronounced in presence of a native microbial community. It is noteworthy that several mycorrhizal helper bacteria release extracellular enzymes favoring the degradation of plant cell walls, helping the fungus to penetrate the root tissues, thereby promoting root colonization [72, 80].
Furthermore, in line with previous studies which showed greater AMF root colonization in co-inoculation treatments [11, 18], here we observed the greatest percentage of AMF root colonization for the B3M, B3HA and, specially, B3HAM treatments compared to inoculation treatments with B3 alone. The extent of root colonization may vary depending on the bacterial inoculant strains and AMF combinations [18, 71]. Bidondo et al. [72] reported some degree of specificity of mycorrhizal helper bacteria with different AMF species, favoring or inhibiting spore germination.
The positive effect of the increased mycorrhizal root colonization on plant performance was underlined by the positive correlation between shoot P content and mycorrhizal root colonization (R = 0.4578, P = 0.0023), emphasizing the benefit to apply appropriate agricultural management that enhances mycorrhizal contribution to crop production.
Microbial community composition
The PLFA analysis and the PCR-DGGE profiles of microbial communities provided interesting insights into treatment dependent shifts. While all inoculation treatments did not significantly affect the total microbial biomass, except for B2, both approaches indicated changes in microbial community composition. Our results showed that the addition of each bacterial inoculant strain, alone or in combination with HA or AMF, induced significant shifts in the structure of bacterial and fungal communities, which were correlated to plant growth promotion and nutrient uptake. The two analytical approaches evidenced consistent results, indicating both that the combination of B. amyloliquefaciens with HA and the mycorrhizal inoculum induced the most extensive change in the soil microbiome compared to the other treatments. Our results confirm recent works that reported shifts in the bacterial community composition in response to the inoculation with Pseudomonas sp., Bacillus amyloliquefaciens and Pseudomonas sp. RU47 strains in tomato plants [64, 81]. Interestingly, we found that Gram+ bacteria increased in inoculated treatments, as also evidenced by the greater values of Gram+/Gram− ratios than for the uninoculated treatments. This is in line with other reports showing that inoculation with Pseudomonas spp, Bacillus amyloliquefaciens and Pseudomonas sp. RU47 increased the relative abundance of the genus Paenibacillus and Bacillus that are classified as Gram+ bacteria and often regarded as plant-beneficial bacteria promoting plant growth [64, 82]. In addition, Gupta et al. [83] reported an increase of four times of Bacillus spp. after inoculation with Bacillus megaterium, suggesting a healthy association of the introduced microorganisms with the native population.
It is also interesting to note that all bacterial inoculants, alone or in combination with HA and AMF, increased the specific marker of arbuscular mycorrhizal fungi and the AMF/saprotrophic fungi ratios more than in the B0 treatment. Furthermore, the growth of AMF, as measured by the specific PLFA marker C16:1ω5, was differentially affected by the presence of the two bacterial strains, in line with previous studies, showing that the identity of microbial strains may significantly affect the AMF spore production and extraradical hyphae growth [18, 72]. Bacillus amyloliquefaciens in combination with HA alone or with mycorrhizal inoculum strongly supported the growth of AMF, more than Pseudomonas spp. Interestingly, even if the mycorrhizal root colonization did not increase significantly in the different B. amyloliquefaciens treatments, instead the abundance of the AMF hyphae in the soil increased significantly in B3HA and B3HAM.
Concomitantly, application of bacterial inoculants impacted negatively the saprotrophic fungal community of soil, as shown in all inoculated treatments by fungal PLFA markers and bacterial/fungi PLFA ratios, which provided smaller and larger values than control, respectively. Cluster analysis of fungal DGGE profiles confirmed these findings. Interestingly, isolates of Paenibacillus have been shown to improve the development of AMF and suppress a broad range of pathogenic fungi [84]. By considering that also P content varied in accordance with the abundance of AMF and saprotrophic fungi, as shown by the positive and negative correlation, respectively (R = 0.7006, P = 0.000; R = − 0.3687, P = 0.0163), these findings suggest that the plant growth promotion and enhanced N and, especially, P uptake could be related to the abundance and activity of indigenous and/or introduced AMF and inhibition of saprotrophic fungi. This agrees with earlier studies showing that the effects of some PGPM may have been due to their interactions with indigenous AMF rather than strictly to their direct induction of plant growth responses [85]. Such mycorrhizal boosting effect is claimed for several microorganisms [11, 19, 72]. Eltbany et al. [64] stated that bacterial inoculants, that stimulate root growth and secretion of phosphatase, may impact soil microbial communities by an increased recruitment of indigenous AMF for the colonization of a larger root system. Similarly, Battini et al. [19] reported that greater P content in shoots of plants inoculated by bacteria was less related to phosphate solubilization than to bacterial induction of an increased root length and/or hyphal abundance in soil. In addition, bacterial inoculants release certain molecules that may inhibit the growth of pathogens detrimental to components of soil microbial community, thereby, indirectly, promoting their growth [84]. Cell wall-degrading enzymes such as β-1,3 glucanase and chitinase are known to be involved in the activity of some P-solubilising microorganisms [86]. Wang et al. [87] isolated two extracellular chitinases from B. amyloliquefaciens that might be important for the antagonistic activity against saprotrophic fungi. These findings may explain, at least partially, the decrease in the population of saprotrophic fungi observed here in the inoculated treatments.
In the present study, the B3HAM treatment, and in general, the treatments where inoculants were combined with HA showed the best plant performances in terms of P uptake, had the greatest root mycorrhizal colonization, AMF/saprotrophic fungi ratios. These findings suggest that HA addition reinforced the positive effects of the bacterial inoculants in promoting AMF development and activity, particularly in combination with B. amyloliquefaciens. Addition of HA may influence processes such as sporulation, production of external mycelium and mycorrhizal colonization [23, 36, 88, 89]. It has been reported that HA rich vermicompost improved AMF root colonization, and this effect was further enhanced in combination with N-fixer bacteria and mycorrhizal inoculation [36]. Moreover, Canellas and Olivares [34] indicated that HS may increase the surface area available for adsorption and establishment of endophytic microorganisms, thus favoring their survival in the soil environment. In fact, inoculated bacteria must overcome complex biotic and abiotic impairing factors to establish a protective niche to survive against competitors and predators [30]. For instance, Meyer et al. [90] reported a loss of more than 99% within 40 days of the inoculated Pseudomonas protegens CHAO cells. On the other hand, Vassilev et al. [87] suggested that production of lytic enzymes by inoculants can be stimulated by the presence of organic substances rich in lignocellulose. Hence, the addition of HA in combination with bioinoculants may have helped to control saprotrophic fungi, favoring competitively AMF population with beneficial effects on plant performance. Moreover, several reports showed that HA modify the composition of microbial communities [36, 91, 92]. In particular, Liu et al. [93], applying amplicon sequencing, reported that HA applications affected both bacterial and fungal communities in soil under maize plants. Interestingly, they showed that HA increased fungal phyla, such as Basidiomycota and Glomeromycota (phylum belonging AMF), thus suggesting that HA enhanced the availability of soil P concentration by increasing the relative abundance of Glomeromycota.
Further studies are required to confirm our results in field conditions and clarify further the specific antagonistic or competitive mechanisms involved in the application of specific bioeffectors. Although our analytical approach provided very informative results, deeper insights could be revealed by applying amplicon sequencing. Furthermore, it could be useful perform concomitantly root and soil metabolomic analysis.
Conclusions and prospects
Our findings revealed that the PGPM bacteria, either alone or in combination with HA and/or AMF, improved plant growth and nutrient uptake in comparison with plants without inoculation. The synergism between Bacillus amyloliquefaciens, HA and mycorrhizal inoculum produced the largest increase in P uptake, while the effect of Pseudomonas spp. combined with humic acids addition was of a lesser extent. Moreover, this positive effect was correlated to the shifts in the microbial community composition. Both techniques used here to assess the microbial community profiles (PLFA and DGGE) showed strong influences of bioeffectors application on soil microbiome. The results evidenced that the shifts in microbial communities positively affected also native AMF community. This was true for all inoculated treatments, but particularly for B3 in combination with HA and mycorrhizal inoculum, showing the largest extraradical mycelium development, as indicated by the largest value of PLFA marker specific for AMF C16:1ω5. These findings indicate the importance of managing the rhizosphere microbiota to emphasize and exploit the potential natural biological fertility of soils. Moreover, the positive effects found here for HA treatments in combination with bacterial strains confirm their potential in boosting the capacity of PGPM to enhance plant performance.
Thus, this study has emphasized the importance of different bioeffectors in improving crop productivity and the critical need to identify the best performing “consortia” or the selection of agricultural management practices favoring microbiota with beneficial functions in different soils and crop species. However, the development of suitable formulations and delivery approaches is extremely important for any field application and acceptance by the growers. These approaches may lead to develop a new generation of biofertilizers for crop production under a bio-sustainable agroecosystem.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
George TS, Hinsinger P, Turner BL. Phosphorus in soils and plants—facing phosphorus scarcity. Plant Soil. 2016;401:1–6.
Arif I, Batool M, Schenk PM. Plant microbiome engineering: expected benefits for improved crop growth and resilience. Trends Biotechnol. 2020;38:1385–96.
King KW, Torbert HA. Nitrate and ammonium losses from surface-applied organic and inorganic fertilizers. J Agric Sci. 2007;145:385–93.
Jansa J, Forczek ST, Rozmoš M, Püschel D, Bukovská P, Hršelová H. Arbuscular mycorrhiza and soil organic nitrogen: network of players and interactions. Chem Biol Technol Agric. 2019;6(1):1–10.
Neumann G, Römheld V. Rhizosphere chemistry in relation to plant nutrition. In: Marschner H, editor. Marschner’s mineral nutrition for higher plants. Cambridge: Academic Press; 2012. p. 347–68.
Frey-Klett P, Garbaye J, Tarkka M. The mycorrhiza helper bacteria revisited. New Phytol. 2007;176:22–36.
Rouphael Y, Franken P, Schneider C, Schwarz D, Giovannetti M, Agnolucci M, et al. Arbuscular mycorrhizal fungi act as biostimulants in horticultural crops. Sci Hort. 2015;196:91–108.
Artursson V, Finlay RD, Jansson JK. Interactions between arbuscular mycorrhizal fungi and bacteria and their potential for stimulating plant growth. Environ Microbiol. 2006;8:1–10.
Owen D, Williams AP, Griffith GW, Withers PJ. Use of commercial bio-inoculants to increase agricultural production through improved phosphorus acquisition. Appl Soil Ecol. 2015;86:41–54.
Berta G, Copetta A, Gamalero E, Bona E, Cesaro P, Scarafoni A, D’Agostino G. Maize development and grain quality are differentially affected by mycorrhizal fungi and a growth-promoting pseudomonad in the field. Mycorrhiza. 2014;24:161–70.
Ghorchiani M, Etesami H, Alikhani HA. Improvement of growth and yield of maize under water stress by co-inoculating an arbuscular mycorrhizal fungus and a plant growth promoting rhizobacterium together with phosphate fertilizers. Agr Ecosyst Environ. 2018;258:59–70.
di Salvioli Fossalunga A, Novero M. To trade in the field: the molecular determinants of arbuscular mycorrhiza nutrient exchange. Chem Biol Technol Agric. 2019;6:1–12.
Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW. Ploughing up the wood-wide web? Nature. 1998;394:431.
Lekberg Y, Koide RT. Is plant performance limited by abundance of arbuscular mycorrhizal fungi? A meta-analysis of studies published between 1988 and 2003. New Phytol. 2005;168:189–204.
Giovannini L, Palla M, Agnolucci M, Avio L, Sbrana C, Turrini A, Giovannetti M. Arbuscular mycorrhizal fungi and associated microbiota as plant biostimulants: research strategies for the selection of the best performing inocula. Agronomy. 2020;10:106.
Singh S, Kapoor KK. Effects of inoculation of phosphate-solubilizing microorganisms and an arbuscular mycorrhizal fungus on mungbean grown under natural soil conditions. Mycorrhiza. 1998;7:249–53.
Roesti D, Gaur R, Johri BN, Imfeld G, Sharma S, Kawaljeet K, Aragno M. Plant growth stage, fertiliser management and bio-inoculation of arbuscular mycorrhizal fungi and plant growth promoting rhizobacteria affect the rhizobacterial community structure in rain-fed wheat fields. Soil Biol Biochem. 2006;38:1111–20.
Ordoñez YM, Fernandez BR, Lara LS, Rodriguez A, Uribe-Vélez D, Sanders IR. Bacteria with phosphate solubilizing capacity alter mycorrhizal fungal growth both inside and outside the root and in the presence of native microbial communities. PLoS ONE. 2016;11:e0154438.
Battini F, Grønlund M, Agnolucci M, Giovannetti M, Jakobsen I. Facilitation of phosphorus, uptake in maize plants by mycorrhizosphere bacteria. Sci Rep. 2017;7:4686.
Nanjundappa A, Bagyaraj DJ, Saxena AK, Kumar M, Chakdar H. Interaction between arbuscular mycorrhizal fungi and Bacillus spp. in soil enhancing growth of crop plants. Fungal Biol Biotechnol. 2019;6:23.
Piccolo A, Nardi S, Concheri G. Structural characteristics of humic substances as related to nitrate uptake and growth regulation in plant systems. Soil Biol Biochem. 1992;24:373–80.
Dobbss LB, Canellas LP, Olivares FL, Aguiar NO, Peres LEP, Azevedo M, et al. Bioactivity of chemically transformed humic matter from vermicompost on plant root growth. J Agric Food Chem. 2010;58:3681–8.
Gryndler M, Hršelová H, Sudová R, Gryndlerová H, Řezáčová V, Merhautová V. Hyphal growth and mycorrhiza formation by the arbuscular mycorrhizal fungus Glomus claroideum BEG 23 is stimulated by humic substances. Mycorrhiza. 2005;15:483–8.
Hršelová H, Soukupová L, Gryndler M. Humic acid-like material from sewage sludge stimulates culture growth of ectomycorrhizal fungi in Vitro. Folia Microbiol. 2007;52:627–30.
Young CC, Rekha P, Lai WA, Arun AB. Encapsulation of plant growth-promoting bacteria in alginate beads enriched with humic acid. Biotechnol Bioeng. 2006;95:76–83.
Piccolo A, Spaccini R, Drosos M, Vinci G, Cozzolino V. The molecular composition of humus carbon: recalcitrance and reactivity in soils. In: Garcia C, Nannipieri P, Hernandez T, editors. The future of soil carbon. Cambridge: Academic Press; 2018. p. 87–124.
Delgado A, Madrid A, Kassem S, Andreu L, del Campillo Md. Phosphorus fertilizer recovery from calcareous soils amended with humic and fulvic acids. Plant Soil. 2002;245:277–86.
Nardi S, Pizzeghello D, Gessa C, Ferrarese L, Trainotti L, Casadoro G. A low molecular weight humic fraction on nitrate uptake and protein synthesis in maize seedlings. Soil Biol Biochem. 2000;32:415–9.
Leme Filho JF, Thomason WE, Evanylo GK, Zhang X, Strickland MS, Chim BK, Diatta AA. The synergistic effects of humic substances and biofertilizers on plant development and microbial activity: a review. Int J Plant Soil Sci. 2020;32:56–75.
Olivares FL, Busato JG, de Paula AM, da Silva LL, Aguiar NO, Canellas LP. Plant growth promoting bacteria and humic substances: crop promotion and mechanisms of action. Chem Biol Technol Agric. 2017;4:30.
Canellas LP, Balmori DM, Médici LO, Aguiar NO, Campostrini E, Rosa RC, et al. A combination of humic substances and Herbaspirillum seropedicae inoculation enhances the growth of maize (Zea mays L.). Plant Soil. 2012;366:119–32.
da Silva SF, Olivares FL, Canellas LP. The biostimulant manufactured using diazotrophic endophytic bacteria and humates is effective to increase sugarcane yield. Chem Biol Technol Agric. 2017;4:24.
da Piedade MA, Olivares FL, Médici LO, Torres-Neto A, Dobbss LB, Canellas LP. Mixed rhizobia and Herbaspirillum seropedicae inoculations with humic acid-like substances improve water-stress recovery in common beans. Chem Biol Technol Agric. 2017;4:1–9.
Canellas LP, Olivares FL. Physiological responses to humic substances as plant growth promoter. Chem Biol Technol Agric. 2014;1:3.
Hansen V, Bonnichsen L, Nunes I, Sexlinger K, Lopez SR, van der Bom FJT, et al. Seed inoculation with Penicillium bilaiae and Bacillus simplex affects the nutrient status of winter wheat. Biol Fertil Soils. 2020;56:97–109.
Maji D, Misra P, Singh S, Kalra A. Humic acid rich vermicompost promotes plant growth by improving microbial community structure of soil as well as root nodulation and mycorrhizal colonization in the roots of Pisum sativum. Appl Soil Ecol. 2017;110:97–108.
Li M, Cozzolino V, Mazzei P, Drosos M, Monda H, Hu Z, Piccolo A. Effects of microbial bioeffectors and P amendements on P forms in a maize cropped soil as evaluated by 31P-NMR spectroscopy. Plant Soil. 2017;427:87–104.
Thonar C, Lekfeldt JDS, Cozzolino V, Kunde D, Kulhánek M, Mosimann C, et al. Potential of three microbial bio-effectors to promote maize growth and nutrient acquisition from alternative phosphorous fertilizers in contrasting soils. Chem Biol Technol Agric. 2017;4:7.
Vinci G, Cozzolino V, Mazzei P, Monda H, Savy D, Drosos M, Piccolo A. Effects of Bacillus amyloliquefaciens and different phosphorus sources on maize plants as revealed by NMR and GC–MS based metabolomics. Plant Soil. 2018;429:437–50.
Vinci G, Cozzolino V, Mazzei P, Monda H, Piccolo A. An alternative to mineral phosphorus fertilizers: the combined effects of Trichoderma harzianum and compost on Zea mays, as revealed by 1H NMR and GC-MS metabolomics. PLoS ONE. 2018;13:e0209664.
Mpanga IK, Nkebiwe PM, Kuhlmann M, Cozzolino V, Piccolo A, Geistlinger J, et al. The form of N supply determines plant growth promotion by P-solubilizing microorganisms in maize. Microorganisms. 2019;7:38.
Bradáčová K, Sittinger M, Tietz K, Neuhäuser B, Kandeler E, Berger N, et al. Maize inoculation with microbial consortia: contrasting effects on rhizosphere activities, nutrient acquisition and early growth in different soils. Microorganisms. 2019;7:329.
Nelson DW, Sommers LE. Total carbon, organic carbon and organic matter. In: Page AL, Miller RH, Keeney DR, editors. Methods of soil analysis, part 2. Madison: American Society of Agronomy; 1982. p. 539–79.
Bremner JM, Mulvaney CS. Nitrogen total. In: Page AL, Miller RH, Keeney DR, editors. Methods of soil analysis, part 2. Madison: American Society of Agronomy; 1982. p. 371–8.
Murphy J, Riley JP. A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta. 1962;27:31–6.
Monda H, Cozzolino V, Vinci G, Drosos M, Savy D, Piccolo A. Molecular composition of the Humeome extracted from different green composts and their biostimulation on early growth of maize. Plant Soil. 2018;429:407–24.
Gericke S, Kurmies B. Die kolorimetrische Phosphorsäure-bestimmung mit AmmoniumVanadat-Molybdat und ihre Anwendung in der Pflanzenanalyse. Z für Pflanzenernähr Düng und Bodenkd. 1952;59:235–47.
Vierheilig H, Coughlan AP, Wyss URS, Piché Y. Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microbiol. 1998;64:5004–7.
Giovannetti M, Mosse B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 1980;84:489–500.
Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–7.
Bardgett RD, Hobbs PJ, Frostegård Å. Changes in soil fungal: bacterial biomass ratios following reductions in the intensity of management of an upland grassland. Biol Fertil Soils. 1996;22:261–4.
Frostegård Å, Bååth E. The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol Fertil Soils. 1996;22:59–65.
Olsson PA. Signature fatty acids provide tools for determination of distribution and interactions of mycorrhizal fungi in soil. Microbiol Ecol. 1999;29:303–10.
Olsson PA, Johansen A. Lipid and fatty acid composition of hyphae and spores of arbuscular mycorrhizal fungi at different growth stages. Mycol Res. 2000;104:429–34.
Sundh I, Nilsson M, Borga P. Variation in microbial community structure in two boreal peatlands as determined by analysis of phospholipid fatty acid profiles. Appl Environ Microbiol. 1997;63:1476–82.
Kroppenstedt RM, Greinermai E, Kornwendisch F. Analysis of fatty-acids and other lipids of actinomycetes and coryneform bacteria. Syst Appl Microbiol. 1984;5:273–273.
Heuer H, Krsek M, Baker P, Smalla K, Wellington EM. Analysis of actinomycetes communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol. 1997;3:3233–41.
Weinert N, Meincke R, Gottwald C, Heuer H, Gomes NCM, Schloter M, et al. Rhizosphere communities of genetically modified zeaxanthin- accumulating potato plants and their parent cultivar differ less than those of different potato cultivars. Appl Environ Microbiol. 2009;75:3859–65.
Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol. 1993;2:113–8.
Heuer H, Wieland G, Schönfeld J, Schönwälder A, Gomes NCM, Smalla K. Bacterial community profiling using DGGE or TGGE analysis. In: Rouchelle P, editor. Environmental molecular microbiology, protocols and applications. Wymondham: Horizon Scientific Press; 2001. p. 177–90.
Rademaker JLW, Louws FJ, Rossbach U, Vinuesa P, de Bruijn FJ. Computer-assisted pattern analysis of electrophoretic fingerprints and database construction. In: Akkermans ADL, van Elsas JD, de Bruijn FJ, editors. Molecular microbial ecology manual. Dordrecht: Academic Publishers; 1999. p. 1–33.
Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, et al. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol. 2001;67:4742–51.
Kropf S, Heuer H, Grüning M, Smalla K. Significance test for comparing complex microbial community fingerprints using pairwise similarity measures. J Microbiol Methods. 2004;57:187–95.
Eltlbany N, Baklawa M, Ding GC, Nassal D, Weber N, Kandeler E, et al. Enhanced tomato plant growth in soil under reduced P supply through microbial inoculants and microbiome shifts. FEMS Microbiol Ecol. 2019;95:fiz124.
Cozzolino V, Di Meo V, Piccolo A. Impact of arbuscular mycorrhizal fungi applications on maize production and soil phosphorus availability. J Geochem Explor. 2013;129:40–4.
Jindo K, Canellas LP, Albacete A, Figueiredo dos Santos L, Frinhani Rocha RL, Baia C, et al. Interaction between humic substances and plant hormones for phosphorous acquisition. Agronomy. 2020;10:640.
Battini F, Cristani C, Giovannetti M, Agnolucci M. Multifunctionality and diversity of culturable bacterial communities strictly associated with spores of the plant beneficial symbiont Rhizophagus intraradices. Microbiol Res. 2016;183:68–79.
Adesemoye AO, Torbert HA, Kloepper JW. Enhanced plant nutrient use efficiency with PGPR and AMF in an integrated nutrient management system. Can J Microbiol. 2008;54:876–86.
Toljander JF, Artursson V, Paul LR, Jansson JK, Finlay RD. Attachment of different soil bacteria to arbuscular mycorrhizal fungal extraradical hyphae is determined by hyphal vitality and fungal species. FEMS Microbiol Let. 2006;254:34–40.
Albertsen A, Ravnskov S, Green H, Jensen DF, Larsen J. Interactions between the external mycelium of the mycorrhizal fungus Glomus intraradices and other soil microorganisms as affected by organic matter. Soil Biol Biochem. 2006;38:1008–14.
Larsen J, Cornejo P, Barea JM. Interactions between the arbuscular mycorrhizal fungus Glomus intraradices and the plant growth promoting rhizobacteria Paenibacillus polymyxa and P. macerans in the mycorrhizosphere of Cucumis sativus. Soil Biol Biochem. 2009;41:286–92.
Bidondo LF, Colombo R, Bompadre J, Benavides M, Scorza V, Silvani V, et al. Cultivable bacteria associated with infective propagules of arbuscular mycorrhizal fungi. Implications for mycorrhizal activity. Appl Soil Ecol. 2016;105:86–90.
Marschner P, Crowley D, Rengel Z. Rhizosphere interactions between microorganisms and plants govern iron and phosphorus acquisition along the root axis—model and research methods. Soil Biol Biochem. 2011;43:883–94.
Rekha PD, Lai WA, Arun AB, Young CC. Effect of free and encapsulated Pseudomonas putida CC–FR2-4 and Bacillus subtilis CC–pg104 on plant growth under gnotobiotic conditions. Bioresour Technol. 2007;98:447–51.
Verma R, Maurya BR, Meena VS. Integrated effect of bio-organics with chemical fertilizer on growth, yield and quality of cabbage (Brassica oleracea var capitata). Indian J Agric Sci. 2014;84:914–9.
Pishchik VN, Vorobyov NI, Walsh OS, Surin VG, Khomyakov YV. Estimation of synergistic effect of humic fertilizer and Bacillus Subtilis on lettuce plants by reflectance measurements. J Plant Nutr. 2016;39:1074–86.
Melo RO, Oliveira HP, Silveira KC, Baldotto LEB, Baldotto MA. Initial performance of maize in response to humic acids and plant growth-promoting bacteria. Rev Ceres. 2018;65:271–7.
Chaiharn M, Lumyong S. Screening and optimization of indole-3-acetic acid production and phosphate solubilization from rhizobacteria aimed at improving plant growth. Curr Microbiol. 2011;62:173–81.
Zhang L, Xu M, Liu Y, Zhang F, Hodge A, Feng G. Carbon and phosphorus exchange may enable cooperation between an arbuscular mycorrhizal fungus and a phosphate-solubilizing bacterium. New Phytol. 2016;210:1022–32.
Lecomte J, St-Arnaud M, Hijri M. Isolation and identification of soil bacteria growing at the expense of arbuscular mycorrhizal fungi. FEMS Microbiol Lett. 2011;317:43–51.
Nassal D, Spohn M, Eltlbany N, Jacquiod S, Smalla K, Marhan S, Kandeler E. Effects of phosphorus-mobilizing bacteria on tomato growth and soil microbial activity. Plant Soil. 2018;427:17–37.
Schreiter S, Sandmann M, Smalla K, Grosch R. Soil type dependent rhizosphere competence and biocontrol of two bacterial inoculant strains and their effects on the rhizosphere microbial community of field-grown lettuce. PLoS ONE. 2014;9:e103726.
Gupta R, Mathimaran N, Wiemken A, Boller T, Bisaria VS, Sharma S. Non-target effects of bioinoculants on rhizospheric microbial communities of Cajanus cajan. Appl Soil Ecol. 2014;76:26–33.
Li B, Ravnskov S, Xie G, Larsen J. Biocontrol of Pythium damping-off in cucumber by arbuscular mycorrhiza-associated bacteria from the genus Paenibacillus. Biocontrol. 2004;52:863–75.
Germida JJ, Walley FL. Plant growth-promoting rhizobacteria alter rooting patterns and arbuscular mycorrhizal fungi colonization of field-grown spring wheat. Biol Fertil Soils. 1996;23:113–20.
Vassilev N, Vassileva M, Nikolaeva I. Simultaneous P-solubilizing and biocontrol activity of microorganisms: potentials and future trends. Appl Microbiol Biotechnol. 2006;71:137–44.
Wang SL, Shih IL, Liang TW, Wang CH. Purification and characterization of two antifungal chitinases extracellularly produced by Bacillus amyloliquefaciens V656 in a shrimp and crab shell powder medium. J Agr Food Chem. 2002;50:2241–8.
Sensoy S, Ocak E, Demi̇r S, Tufenkci̇ S. Effects of humic acid, whey and Arbuscular Mycorrhizal Fungi (AMF) applications on seedling growth and Fusarium wilt in zucchini (Cucurbita pepo L.). J Anim Plant Sci. 2013;23:507–13.
Pinos NQ, Berbara RLL, Sanchez SE, van Tol de Castro TA, García AC. Combination of humic substances and arbuscular mycorrhizal fungi affecting corn plant growth. J Environ Qual. 2019;48:1594–604.
Meyer G, Bünemann EK, Frossard E, Maurhofer M, Mäder P, Oberson A. Gross phosphorus fluxes in a calcareous soil inoculated with Pseudomonas protegens CHA0 revealed by 33P isotopic dilution. Soil Biol Biochem. 2017;104:81–94.
Puglisi E, Pascazio S, Suciu N, Cattani I, Fait G, Spaccini R, et al. Rhizosphere microbial diversity as influenced by humic substance amendments and chemical composition of rhizodeposits. J Geochem Explor. 2013;129:82–94.
Schoebitz M, López MD, Serrí H, Martínez O, Zagal E. Combined application of microbial consortium and humic substances to improve the growth performance of blueberry seedlings. J Soil Sci Plant Nutr. 2016;16:1010–23.
Liu M, Wang C, Wang F, Xie Y. Maize (Zea mays) growth and nutrient uptake following integrated improvement of vermicompost and humic acid fertilizer on coastal saline soil. Appl Soil Ecol. 2019;142:147–54.
Acknowledgements
Not applicable.
Funding
This work was supported by the European Community’s Seventh Framework Program 662 (FP7/2007–2013) under the Grant Agreement No. 312117 (BIOFECTOR).
Author information
Authors and Affiliations
Contributions
VC and HM conceived the study and set up the experiments, analyzed data and drafted the manuscript. HM performed extraction of humic acids, plant analyses and microbiological experiments. VC evaluated the percentage of mycorrhizal colonization and revised the final manuscript. DS, GV, and VDM assisted in setting up the experiments, plant and soil sample collection and their preparation for analyses. KS supervised microbiological analyses and revised the draft version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent in participate
This manuscript is an original paper and has not been published in other journals. The authors agreed to keep the copyright rule.
Consent for publication
The authors agreed to the publication of the manuscript in this journal.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file1
: Figure S1 DGGE fingerprints of bacterial 16S rRNA gene fragments from total community DNA. Figure S2 Fungal DGGE fingerprints of ITS fragments.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Cozzolino, V., Monda, H., Savy, D. et al. Cooperation among phosphate-solubilizing bacteria, humic acids and arbuscular mycorrhizal fungi induces soil microbiome shifts and enhances plant nutrient uptake. Chem. Biol. Technol. Agric. 8, 31 (2021). https://doi.org/10.1186/s40538-021-00230-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40538-021-00230-x