Abstract
Background
Anopheles sinensis (Diptera: Culicidae) is a primary vector of Plasmodium vivax and Brugia malayi in most regions of China. In addition, its phylogenetic relationship with the cryptic species of the Hyrcanus Group is complex and remains unresolved. Mitochondrial genome sequences are widely used as molecular markers for phylogenetic studies of mosquito species complexes, of which mitochondrial genome data of An. sinensis is not available.
Methods
An. sinensis samples was collected from Shandong, China, and identified by molecular marker. Genomic DNA was extracted, followed by the Illumina sequencing. Two complete mitochondrial genomes were assembled and annotated using the mitochondrial genome of An. gambiae as reference. The mitochondrial genomes sequences of the 28 known Anopheles species were aligned and reconstructed phylogenetic tree by Maximum Likelihood (ML) method.
Findings
The length of complete mitochondrial genomes of An. sinensis was 15,076 bp and 15,138 bp, consisting of 13 protein-coding genes, 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and an AT-rich control region. As in other insects, most mitochondrial genes are encoded on the J strand, except for ND5, ND4, ND4L, ND1, two rRNA and eight tRNA genes, which are encoded on the N strand. The bootstrap value was set as 1000 in ML analyses. The topologies restored phylogenetic affinity within subfamily Anophelinae. The ML tree showed four major clades, corresponding to the subgenera Cellia, Anopheles, Nyssorhynchus and Kerteszia of the genus Anopheles.
Conclusions
The complete mitochondrial genomes of An. sinensis were obtained. The number, order and transcription direction of An. sinensis mitochondrial genes were the same as in other species of family Culicidae.
Similar content being viewed by others
Multilingual abstracts
Please see Additional file 1 for translations of the abstract into the six official working languages of the United States.
Background
Anopheles sinensis Wiedemann, 1828 is an oriental species with a wide distribution in China [1]. It is a vector of Plasmodium vivax in plain regions of central China, and certain worms such as Brugia malayi that cause lymphatic filariasis [2, 3]. Despite its disputable malaria vector capacity, An. sinensis is still incriminated as a competent vector for Plasmodium vivax malaria due to its abundant population size and wide distribution, which have led to occasional local malaria epidemics or outbreaks throughout history [4].
Anopheles sinensis is one of the members in the Hyrcanus Group. The Hyrcanus Group is an extremely complex species assemblage of the genus Anopheles subgenus Anopheles, which includes above 20 closely related species in China [1, 5, 6]. Because of similar morphological characters of female adult, the identification of these species in the group has been taxonomically problematic. Such as, An. sinensis was almost impossible distinguished from its sibling species (An. lesteri, An. yatsushiroensis, An. kleini, and so on) [1]. The PCR assay was established by sequences of second internal transcribed spacer (ITS2) region of the ribosomal DNA (rDNA) to identify An. sinensis from its cryptic species members of Hyrcanus Group [5]. The genetic structure of An. sinensis populations in China were also detected by molecular markers, and the weak genetic structure may be a consequence of low genetic differentiation and high gene flow among populations in central China [7,8,9,10]. However, there are still some issues to be elucidated in the molecular classification of An. sinensis, such as natural hybrid between An. kleini and An. sinensis was discovered in the Republic of Korea [11, 12] and China (unpublished data). So, the evolutionary relationship of An. sinensis, such as speciation and other issues need to be elucidated further.
Mitochondrial genomes strictly follow maternal inheritance in structure and evolution, and contain abundant information for population genetic and phylogenetic studies [13,14,15,16,17,18,19,20,21]. There is no adequate mitogenome information available for An. sinensis. In this study, we used next-generation sequencing to characterize the mitochondrial genomes of An. sinensis, and to reconstruct phylogenetic tree of the known Anopheles species.
Methods
Mosquito collection and species identification
Wild mosquito adults were collected by CDC mini light traps (BioQuip, USA) or artificial catching aspirator at livestock corrals from Jining and Caoxian County in Shandong Province, China in July, 2012. With the owners’ consent, the light traps were set up in cow pens from 18:30 pm to 8:30 am next day. Mosquitoes of the An. hyrcanus group were sorted out in the field by morphology using the identification keys [1], and kept individually in silica gel filled tubes at 4 °C until DNA extraction. After being brought back to the laboratory, the female adults were separated into head and body. The single head was used to identify species by PCR assay based on rDNA ITS2 sequences [5]. Twenty bodies pool of An. sinensis species was extracted genomic DNA using Meta-G-Nome™ DNA Isolation Kit (Epicentre, USA), followed by Illumina sequencing.
Metagenome sequencing
Genomic DNA was fragmented and sequencing libraries were prepared, which insert size was about 700 bp. Double-end pairing sequencing was performed using Illumina HiSeq 2000 (Genewiz, USA). Each library was sequenced to generate about 20 million paired-end reads from each sample. The mean length of reads was 101 base pairs.
Mitochondrial genome assembly and annotation
The quality of total reads was controlled by FASTQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), then the high quality reads were mapping to An. sinensis genome (SAMN02910229) [22]. The contigs were assembled by KmerGenie softwares [23], and aligned with mitochondrial genome of Culicidae mosquitoes on GenBank BLAST website, which identity threshold value was set as 90%. The complete mitochondrial genome of An. sinenesis was obtained, including 13 protein coding genes, 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes and AT-rich control region. The genes and region of the mitochondrial genome were identified by comparison with the reference mitogenome sequences of An. gambiae (GenBank Accession No. L20934.1) and other Anopheles mosquitoes. Some parts fragments such as CO1 and CO2 region were verified by PCR products [7, 24].
Phylogenetic analysis
A total of 28 Anopheles species (including An. sinensis in this study) mitochondrial genome sequences from NCBI database available were analyzed by MEGA 6 software [25]. The phylogenetic relationships of the mitogenome DNA sequences for Anopheles mosquitoes were reconstructed using Maximum Likelihood (ML) method with PhyML 3.0 [26]. The best fit model of nucleotide substitution, the GTR + I + G model, was determined for the ML tree inference with Modeltest 3.7 [27]. The bootstrap values for 1000 replicates were calculated.
Results and discussion
Mitogenome organization and composition
The length of two complete mitochondrial genomes of An. sinensis were obtained from two samples, which were 15,076 bp and 15,138 bp, respectively. Both sequences was conserved, except the length of AT-rich control region. One of the complete mitochondrial genome sequence was chosen for further analysis, which was submitted to GenBank (Accession No. KT218684.1).
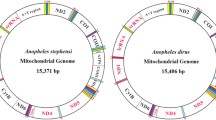
The mitochondrial genomes of An. sinensis mosquitoes consisted of circular DNA molecules, contained 13 protein-coding genes, 22 transfer RNA (tRNA) genes, 2 rRNA genes (12S rRNA and 16S rRNA), and an AT-rich control region (Fig. 1). The mitochondrial genes showed no length variation on either the J or N strand. The AT-rich control region was located between the SrRNA and tRNA-Ile genes.
Graphic representation of the gene arrangement and gene order of the mitochondrial genome of An. sinensis. All 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, and the AT-rich control region are indicated in the circle. Each tRNA gene is identified by its single letter abbreviation. The direction of transcription is indicated by colour, the genes in blue content (J strand) as clockwise and in green (N strand) as counterclockwise
Protein coding genes
The mitochondrial genome of An. sinensis species consisted of 13 intron-less protein coding genes. The gene number, order and transcription direction of mitochondrial genes were the same as in other species of Culicidae species [13,14,15, 17, 19, 21, 28,29,30,31,32,33]. There were 9 genes located in the J strand, as ND2, CO1, CO2, ATPase8, ATPase6, CO3, ND3, ND6 and CytB, while ND1, ND5, ND4 and ND4L located in the N strand. The length of nucleotides and amino acids of these 13 genes were showed in Table 1.
Transfer and ribosomal RNAs genes
All 22 tRNA genes were dispersed in the mitochondrial genome of An. sinensis, the length and position of them was similar with the reported Culicidae mosquitoes (Table 2) [13,14,15, 20, 21, 28, 29, 31,32,33]. Among them, there were two kinds of transfer genes for serine and leucine, and the remaining 18 tRNA genes correspond to the other amino acid respectively. The arrangement of tRNA-Arg and tRNA-Ala was inverse compared with the insects of family Drosophilidae, Tephritidae, Calliphoridae, Muscidae and Ceratopogonidae [34,35,36].
There were 2 rRNA genes in mitochondrial genome of An. sinensis, both located in the N strand. The length of SrRNA gene was 692 bp encoding 12S rRNA, and the LrRNA gene was 1329 bp encoding 16S rRNA.
Phylogenetic analysis
The molecular phylogenetic relationship using complete mitochondrial genome sequence of 28 Anopheles species was analyzed. All sequences generated in this study have been deposited in the GenBank. The topologies restored phylogenetic affinity within subfamily Anophelinae (Fig. 2). The ML tree showed that two major clades with mosquitoes in subgenus Kerteszia (n = 4) and the other three subgenera, as Cellia (n = 16), Anopheles (n = 3), Nyssorhynchus (n = 5) in genus Anopheles. The latter subgenera Cellia, Anopheles and Nyssorhynchus were monophyly, and the subgenera Cellia with Anopheles was sister relationship. The bootstrap values were almost above 50%.
Phylogenetic tree generated using the Maximum Likelihood method based on complete mitochondrial genomes. The bootstrap values are marked on each node of the tree. The GenBank Accession No. is in bracket: An. sinensis (KT218684.1), An. arabiensis (KT382816), An. quadrimaculatus A (L04272.1), An. darlingi (GQ918272.1), An. atroparvus (KT382817.1), An. minimus (KT895423.1), An. dirus A (JX219731.1), An. coluzzii (KT382819.1), An. oryzalimnetes (HQ335345.1), An. albitarsis (HQ335344.1), An. culicifacies B (KR732656.1), An. epiroticus (KT382821.1), An. gambiae (L20934.1), An. farauti (JX219741.1), An. deaneorum (HQ335347.1), An. merus (KT382824.1), An. melas (KT382823.1), An. janconnae (HQ335348.1), An. hinesorum (JX219734.1), An. cracens (JX219733.1), An. maculatus (KT382822.1), An. stephensi (KT382827.1), An. punctulatus (JX219744.1), An. christyi (KT382818.1), An. cruzii (KJ701506.1), An. laneanus (KU551288.1), An. homunculus (KU551283.1), An. bellator (KU551287.1)
Conclusions
The complete mitochondrial genomes of An. sinensis were obtained. The number, order and transcription direction of An. sinensis mitochondrial genes were the same as in other species of Culicidae. And the complete mitogenome data can provide basic information for analyzing phylogenetic relationship of mosquito species.
Abbreviations
- ITS2:
-
Second internal transcribed spacer
- ML:
-
Maximum likelihood
- rDNA:
-
Ribosomal DNA
- rRNA:
-
Ribosomal RNA
- tRNA:
-
Transfer RNA
References
Lu B. Fauna Sinica, Insecta Vol. 9: Diptera, Culicidae II. Beijing: Science Press; 1997.
Zhang SQ, Zhang QJ, Cheng F, Wang LL, Pen GP. Threshold of transmission of Brugia malayi by Anopheles sinensis. J Trop Med Hyg. 1991;94(4):245–50.
Zhou SS, Zhang SS, Wang JJ, Zheng X, Huang F, Li WD, et al. Spatial correlation between malaria cases and water-bodies in Anopheles sinensis dominated areas of Huang-Huai plain, China. Parasit Vectors. 2012;5:106. doi:10.1186/1756-3305-5-106.
Ren Z, Wang D, Hwang J, Bennett A, Sturrock HJ, Ma A, et al. Spatial-temporal variation and primary ecological drivers of Anopheles sinensis human biting rates in malaria epidemic-prone regions of China. PLoS One. 2015;10(1):e0116932. doi:10.1371/journal.pone.0116932.
Ma Y, Xu J. The Hyrcanus group of Anopheles (Anopheles) in China (Diptera: Culicidae): species discrimination and phylogenetic relationships inferred by ribosomal DNA internal transcribed spacer 2 sequences. J Med Entomol. 2005;42(4):610–9.
Ma Y, Xu J. Progress of taxonomic study on the Anopheline mosquitoes in China. Chin J Vector Biol Control. 2015;26(5):433–8. (in Chinese)
Feng X, Huang L, Lin L, Yang M, Ma Y. Genetic diversity and population structure of the primary malaria vector Anopheles sinensis (Diptera: Culicidae) in China inferred by cox1 gene. Parasit Vectors. 2017;10(1):75. doi:10.1186/s13071-017-2013-z.
Chang X, Zhong D, Lo E, Fang Q, Bonizzoni M, Wang X, et al. Landscape genetic structure and evolutionary genetics of insecticide resistance gene mutations in Anopheles sinensis. Parasit Vectors. 2016;9:228. doi:10.1186/s13071-016-1513-6.
Ma Y, Yang M, Fan Y, Wu J, Xu J. Population structure of the malaria vector Anopheles sinensis (Diptera: Culicidae) in China: two gene pools inferred by microsatellites. PLoS One. 2011;6(7):e22219. doi:10.1371/journal.pone.0022219.
Makhawi AM, Liu XB, Yang SR, Liu QY. Genetic variations of ND5 gene of mtDNA in populations of Anopheles sinensis (Diptera: Culicidae) malaria vector in China. Parasit Vectors. 2013;6:290. doi:10.1186/1756-3305-6-290.
Choochote W, Min GS, Intapan PM, Tantrawatpan C, Saeung A, Lulitanond V. Evidence to support natural hybridization between Anopheles sinensis and Anopheles kleini (Diptera: Culicidae): possibly a significant mechanism for gene introgression in sympatric populations. Parasit Vectors. 2014;7:36. doi:10.1186/1756-3305-7-36.
Joshi D, Choochote W, Min GS. Short report: Natural hybrid between Anopheles kleini and Anopheles sinensis. Am J Trop Med Hyg. 2009;81(6):1020–2. doi:10.4269/ajtmh.2009.09-0123.
Oliveira TM, Foster PG, Bergo ES, Nagaki SS, Sanabani SS, Marinotti O, et al. Mitochondrial genomes of Anopheles (Kerteszia) (Diptera: Culicidae) from the Atlantic forest, Brazil. J Med Entomol. 2016;53(4):790–7.
Luo QC, Hao YJ, Meng F, Li TJ, Ding YR, Hua YQ, et al. The mitochondrial genomes of Culex tritaeniorhynchus and Culex pipiens pallens (Diptera: Culicidae) and comparison analysis with two other Culex species. Parasit Vectors. 2016;9(1):406. doi:10.1186/s13071-016-1694-z.
Hua YQ, Ding YR, Yan ZT, Si FL, Luo QC, Chen B. The complete mitochondrial genome of Anopheles minimus (Diptera: Culicidae) and the phylogenetics of known Anopheles mitogenomes. Insect Sci. 2016;23(3):353–65. doi:10.1111/1744-7917.12326.
Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phylogenet Evol. 2006;39(2):417–23. doi:10.1016/j.ympev.2006.01.006.
Moreno M, Marinotti O, Krzywinski J, Tadei WP, James AA, Achee NL, et al. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar J. 2010;9:127. doi:10.1186/1475-2875-9-127.
Krzywinski J, Li C, Morris M, Conn JE, Lima JB, Povoa MM, et al. Analysis of the evolutionary forces shaping mitochondrial genomes of a Neotropical malaria vector complex. Mol Phylogenet Evol. 2011;58(3):469–77. doi:10.1016/j.ympev.2011.01.003.
Logue K, Chan ER, Phipps T, Small ST, Reimer L, Henry-Halldin C, et al. Mitochondrial genome sequences reveal deep divergences among Anopheles punctulatus sibling species in Papua New Guinea. Malar J. 2013;12:64. https://doi.org/10.1186/1475-2875-12-64.
Chu H, Li C, Guo X, Zhang H, Luo P, Wu Z, et al. The phylogenetic relationships of known mosquito (Diptera: Culicidae) mitogenomes. Mitochondrial DNA A DNA Mapp Seq Anal. 2016:1–5. doi:10.1080/24701394.2016.1233533.
Demari-Silva B, Foster PG, de Oliveira TM, Bergo ES, Sanabani SS, Pessoa R, et al. Mitochondrial genomes and comparative analyses of Culex camposi, Culex coronator, Culex usquatus and Culex usquatissimus (Diptera:Culicidae), members of the coronator group. BMC Genomics. 2015;16:831. doi:10.1186/s12864-015-1951-0.
Zhou D, Zhang D, Ding G, Shi L, Hou Q, Ye Y, et al. Genome sequence of Anopheles sinensis provides insight into genetics basis of mosquito competence for malaria parasites. BMC Genomics. 2014;15:42. doi:10.1186/1471-2164-15-42.
Chikhi R, Medvedev P. Informed and automated k-mer size selection for genome assembly. Bioinformatics. 2014;30(1):31–7. doi:10.1093/bioinformatics/btt310.
Ma Y, Ma Y, Wang Y. Phylogenetic relationship of Anopheles species, subgenus Anopheles in China: based on mtDNA and rDNA sequences. China Trop Med. 2015;15(11):1281–8. (in Chinese)
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. doi:10.1093/molbev/mst197.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21. doi:10.1093/sysbio/syq010.
Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14(9):817–8.
Beard CB, Hamm DM, Collins FH. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 1993;2(2):103–24.
Mitchell SE, Cockburn AF, Seawright JA. The mitochondrial genome of Anopheles quadrimaculatus species A: complete nucleotide sequence and gene organization. Genome. 1993;36(6):1058–73.
Behura SK, Lobo NF, Haas B, de Bruyn B, Lovin DD, Shumway MF, et al. Complete sequences of mitochondria genomes of Aedes aegypti and Culex quinquefasciatus and comparative analysis of mitochondrial DNA fragments inserted in the nuclear genomes. Insect Biochem Mol Biol. 2011;41(10):770–7. doi:10.1016/j.ibmb.2011.05.006.
Hardy CM, Court LN, Morgan MJ. The complete mitochondrial DNA genome of Aedes vigilax (Diptera: Culicidae). Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(4):2552–3. doi:10.3109/19401736.2015.1038800.
Hua YQ, Yan ZT, Fu WB, He QY, Zhou Y, Chen B. Sequencing and analysis of the complete mitochondrial genome in Anopheles culicifacies species B (Diptera: Culicidae). Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(4):2909–10. doi:10.3109/19401736.2015.1060434.
Zhang H, Xing D, Wang G, Li C, Zhao T. Sequencing and analysis of the complete mitochondrial genome of Aedes albopictus (Diptera: Culicidae) in China. Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(4):2787–8. doi:10.3109/19401736.2015.1053067.
Clary DO, Wahleithner JA, Wolstenholme DR. Transfer RNA genes in Drosophila mitochondrial DNA: related 5′ flanking sequences and comparisons to mammalian mitochondrial tRNA genes. Nucleic Acids Res. 1983;11(8):2411–25.
Li X, Wang Y, Su S, Yang D. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA A DNA Mapp Seq Anal. 2016;27(2):1435–6. doi:10.3109/19401736.2014.953080.
Spanos L, Koutroumbas G, Kotsyfakis M, Louis C. The mitochondrial genome of the mediterranean fruit fly, Ceratitis capitata. Insect Mol Biol. 2000;9(2):139–44.
Acknowledgements
We are grateful to Mao-Qing Gong and Peng Cheng (Shandong Provincial Institute of Parasitic diseases control), Chun-Yan Nan, Tong Wu, who provided field assistance in the study.
Funding
This work was supported by the National Natural Sciences Foundation of China (No. 81371848) and the Infective Diseases Prevention and Cure Project of China (No. 2017ZX10303404–002).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Author information
Authors and Affiliations
Contributions
All authors read and approved the final version of the manuscript. YM designed the study and did morphological identification. YM and XL collected mosquito specimens in the field, YW did species molecular identification. KC, YW and HP did data analysis. KC, YW, HP and YM wrote the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
No specific permits were required for this study. The study did not involve endangered or protected species. Therefore, the local ethics committee deemed that approval was unnecessary.
Consent for publication
Not applicable.
Competing interests
The authors declared that they have no competing interests.
Additional file
Additional file 1:
Multilingual abstracts in the six official working languages of the United Nations. (PDF 666 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chen, K., Wang, Y., Li, XY. et al. Sequencing and analysis of the complete mitochondrial genome in Anopheles sinensis (Diptera: Culicidae). Infect Dis Poverty 6, 149 (2017). https://doi.org/10.1186/s40249-017-0362-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40249-017-0362-7