Abstract
Background
Members of the Anopheles punctulatus group (AP group) are the primary vectors of human malaria in Papua New Guinea. The AP group includes 13 sibling species, most of them morphologically indistinguishable. Understanding why only certain species are able to transmit malaria requires a better comprehension of their evolutionary history. In particular, understanding relationships and divergence times among Anopheles species may enable assessing how malaria-related traits (e.g. blood feeding behaviours, vector competence) have evolved.
Methods
DNA sequences of 14 mitochondrial (mt) genomes from five AP sibling species and two species of the Anopheles dirus complex of Southeast Asia were sequenced. DNA sequences from all concatenated protein coding genes (10,770 bp) were then analysed using a Bayesian approach to reconstruct phylogenetic relationships and date the divergence of the AP sibling species.
Results
Phylogenetic reconstruction using the concatenated DNA sequence of all mitochondrial protein coding genes indicates that the ancestors of the AP group arrived in Papua New Guinea 25 to 54 million years ago and rapidly diverged to form the current sibling species.
Conclusion
Through evaluation of newly described mt genome sequences, this study has revealed a divergence among members of the AP group in Papua New Guinea that would significantly predate the arrival of humans in this region, 50 thousand years ago. The divergence observed among the mtDNA sequences studied here may have resulted from reproductive isolation during historical changes in sea-level through glacial minima and maxima. This leads to a hypothesis that the AP sibling species have evolved independently for potentially thousands of generations. This suggests that the evolution of many phenotypes, such as insecticide resistance will arise independently in each of the AP sibling species studied here.
Similar content being viewed by others
Background
Anopheles mosquitoes are distributed worldwide, with the exception of Antarctica, and feed on a variety of hosts from birds to mammals ([1] and references therein). Within the Anopheles genus, 70 of over 500 species are able to transmit human malaria[2]. These include the well-known species Anopheles gambiae, Anopheles arabiensis and Anopheles funestus that are the main vectors of malaria in Africa, Anopheles darlingi and Anopheles albitarsis in South America, Anopheles dirus and Anopheles minimus in Southeast Asia (SEA) and Anopheles punctulatus in Southwest Pacific (SW Pacific).
Despite their medical importance, the current understanding of the Anopheles phylogeny – the relationship among species as well as the times they diverged from each other – remains limited. Studies of this nature are complicated in Anopheles by the existence of species complexes (including morphologically identical sibling species)[3–6] and incipient species[7–9], as well as by the paucity of genetic data for most species besides An. gambiae. The current hypothesis of Anopheles evolution, mostly based on the extant geographic distribution of Anopheles mosquitoes, proposes that Anopheles originated in western Gondwana during the Cretaceous. They subsequently migrated across the world aided by land connections and radiated into multiple species adapted to unique habitats and climatic conditions[10, 11]. However, the timing and routes of these dispersions are unknown and the relationships among current species remain poorly understood. Determining the evolutionary relationships among Anopheles species has important clinical and vector control implications as it could clarify whether traits required for transmission of human blood-borne pathogens, avoidance of long-lasting insecticide-treated nets or insecticide resistance evolved only once in an ancestral population or, alternatively, whether different species acquired these traits independently.
The focus of this study is on members of the Anopheles punctulatus (AP) group, the principal vectors of malaria in Papua New Guinea (PNG), the Solomon Islands, Vanuatu, and northern Australia[12, 13]. Historically, the AP group was separated into four species based on morphological differences in the proboscis and wings: An. punctulatus, Anopheles koliensis, Anopheles farauti, and Anopheles clowi[12]. Later studies involving cross-mating[14–16], allozyme analyses[17] and DNA sequence analysis[18–22] provided evidence suggesting further differentiation of the AP group into 13 species, most of them morphologically indistinguishable. At least five of these species - An. punctulatus s.s, An. koliensis, An. farauti s.s (previously An. farauti 1), Anopheles hinesorum (previously An. farauti 2), and An. farauti 4 - have been described as competent vectors of malaria[23, 24]. Phylogenetic studies of this group have focused on DNA sequences of ribosomal RNAs[25, 26], mitochondrial genes[27, 28], ribosomal ITS2[19], and voltage-gated sodium channel gene[29]. However, the genetic information generated in these studies has not allowed robust determination of the AP group phylogeny and has often yielded conflicting results[19, 27]. In addition, no study has yet evaluated the relationships between AP sibling species and other Anopheles species from neighbouring regions, such as species from SEA. As Beebe and Cooper have described this group of Anopheles sibling species as ‘unspecialized’, there is some reason to hypothesize that AP group members have evolved to acquire a broad range of feeding strategies to ensure survival[30].
Here, the evolutionary history of the AP group of PNG is investigated by sequencing the mitochondrial (mt) genome of 14 individual mosquitoes from the AP group and neighbouring An. dirus complex. Next generation sequencing technologies were used to generate mt genome sequences and de novo assemble each individual sequence. The concatenated sequence of all mitochondrial protein coding genes (10,770 bp) was used to reconstruct robust phylogenies and estimate divergence times among available Anopheles mt genomes. The implications of this study’s findings are discussed with regard to the evolutionary history of anophelines in general and the origin of the AP group.
Methods
Sampling and laboratory procedures
Sample collection and DNA extraction
AP mosquitoes were obtained from the Entomology Unit of the PNG Institute of Medical Research (PNGIMR). Anopheles dirus samples were obtained from the Faculty of Tropical Medicine, Mahidol University (Thailand). Genomic DNA from individual mosquitoes was extracted using the Qiagen DNeasy® blood and tissue kit according to the supplementary protocol for purification of insect DNA. The species of each AP mosquito was determined using a PCR-based assay targeting the ITS2 locus[31].
Whole genome shotgun sequencing and assembly
The whole genome of five mosquitoes (An. punctulatus s.s. (n = 1), An. koliensis (n = 1), An. farauti s.s. (n = 1), and An. farauti 4 (n = 2)) were sequenced. Genomic DNA was fragmented into ~300 base pairs and sequencing libraries were prepared using the New England Biolabs (NEB) NEBNext® kit protocol and standard Illumina paired-end adaptors. Each library was sequenced on one lane of an Illumina GAIIx or HiSeq2000 instrument to generate 37 to 150 million paired-end reads from each sample [see Additional file1].
To identify reads originating from the mt genome and separate them from reads originating from the nuclear genome, the program Bowtie[32] was used to map all reads generated from one sample on the cytochrome oxidase I (COI), cytochrome oxidase II (CO2), and the voltage gated sodium channel (VGSC) gene sequences previously generated for each species. As expected based on the copy number, the sequence coverage of mt genes, COI and CO2, was 50–60 fold greater than the coverage of the nuclear gene, VGSC [see Additional file2]. The ~500 X coverage of mtDNA implied that multiple identical reads mapped to the exact same nucleotide position along the entire mt genome sequence, therefore, One hypothesis is that most reads occurring twice or more in each shotgun sequencing dataset were likely to originate from the mt genome. These reads were therefore selected (regardless of their DNA sequence) for reconstructing the complete mtDNA sequence of each sample. All assemblies were performed using ABySS[33] with a k-mer size of 29 and C = 70. The assembled contigs were aligned to the mtDNA sequence of An. gambiae with MUMmer[34] and any remaining gaps were filled by PCR and Sanger sequencing [see Additional file3]. To identify possible artefacts or assembly errors, all reads generated from a given sample were then mapped to the final mtDNA contig (assembled from a subset of these reads). If necessary, any base in the contig differing from the nucleotide carried by a majority of the reads was replaced by this nucleotide.
Multiplexed mt genome sequencing and assembly
For the remaining nine samples, a multiplex approach was used to simultaneously sequence the mt genome [see Additional file4]. First, primers were designed to amplify any Anopheles mt genome using seven overlapping long-range PCRs. An Anopheles consensus sequence was generated by aligning the mt genomes of An. punctulatus s.s., An. farauti s.s, An. gambiae[35], An. quadrimaculatus[36] and An. darlingi[37] with ClustalW[38] and masking any variant site. Primers were then designed based on this consensus sequence using Primer3[39] following the Roche Expand Long Range dNTPack® kit recommendations. Primers were able to be designed at overlapping sites with two or less variants and without known variants in the last 3’ positions [see Additional file3].
Each amplicon was amplified using the Roche Expand Long Range dNTPack® kit protocol with 20–40 ng of gDNA per PCR reaction and 3% DMSO. Amplification conditions were as follows: 3 minute denaturation step at 94°C, 39 cycles of 94°C for 45 seconds, 50°C for 45 seconds, 60°C for 5 minutes followed by a 10 minute final elongation at 60°C. Product amplification was verified by electrophoresis on a 1% agarose gel.
Following amplification, all seven PCR products from a given individual were pooled together and DNA molecules were sheared into 300 bp fragments [see Additional file4]. A sequencing library for each individual was prepared using Illumina adapters including a unique 6-nucleotide barcode. Finally, libraries were pooled from all mosquitoes in equal concentrations and the resulting pool was sequenced on one lane of an Illumina HiSeq 2000 instrument [Additional file4], generating an average of 28 million paired-end reads of 100 bp per sample [see Additional file1] [NCBI SRA: SRP013853].
The mtDNA sequence of each sample was independently assembled. To normalize coverage along the mt genome (necessary to de novo assemble the DNA sequence) and remove reads containing sequencing errors, the number of reads carrying each DNA sequence observed was calculated. Given the very high sequence coverage, DNA sequences observed rarely likely correspond to reads with sequencing error(s). Therefore, only DNA sequences represented by >20 reads were used to de novo assemble each mt genome. In addition, to normalize coverage and facilitate computation, only one instance of each such sequence was used. This subset of reads was used for de novo assembly of the mt genomes using ABySS with a k-mer size of 31. The resulting contigs were aligned to the mtDNA of An. gambiae using MUMmer. Overlapping contigs were collapsed using nucleotide ambiguity codes in the SeqMan Pro™ program in DNASTAR’s LASERGENE® Core Suite[40] to produce a consensus sequence for each sample. All reads (i.e., not only the subset of reads used to generate the assembly) generated from a given sample were then mapped to its consensus sequence using bwa[41] and the nucleotide determination at each position of the genome sequence was validated using the Samtool mpileup[42] and perl scripts. All mt genome sequences have been deposited in GenBank (see Table 1 for accession numbers). In addition, all complete Anopheles mt genomes available from NCBI were retrieved[35–37, 43] as well as mt genomes from Aedes [GenBank:EU352212.1 and GenBank:AY072044.1], Culex [GenBank:HQ724614.1] and Drosophila[44, 45] (Table 1). Note that six of the 15 mt genomes downloaded from NCBI belong to sibling species of the An. albitarsis group from South America (An. albitarsis, An. albitarsis F, An. albitarsis G, Anopheles deaneorum, Anopheles janconnae and Anopheles oryzalimnetes).
Data analysis
Phylogenetic analysis and molecular dating Since relying on the gene annotations from one of the Anopheles (i.e. An. gambiae) may introduce systematic biases in the phylogenetic analyses, the DNA sequence of each gene for each mt genome (i.e. the sequences that were generated as well as those retrieved from NCBI) was determined with tBlastn using the Drosophila melanogaster protein annotations as references [Genbank:U37541.1]. Briefly, for each protein coding sequence, the DNA sequences were translated into amino acid sequences, aligned to each other and the amino acid sequence was reverse-translated back into nucleotide sequences with Translator X[46] using the default parameters and the invertebrate mt genetic code. The aligned coding protein sequences from all 13 mt genes (resulting in 10,770 nucleotides) were concatenated and the best model of nucleotide substitutions was determined using the program jModeltest v0.1.1[47]. According to the Akaike Information Criterion, the best nucleotide substitution model for this data set was the General Time Reversible with gamma distribution (GTR + G) model.
Bayesian phylogenies were reconstructed using BEAST v1.7.2[48] with the following program parameters: an uncorrelated lognormal relaxed clock model, allowing for rate heterogeneity among species, the GTR + G substitution model, the SRD06 model of partitioning, which allows estimation of nucleotide substitution parameters separately for the 1st + 2nd and 3rd codon positions, and a Yule model for tree reconstruction. Using the above parameters, three independent runs of 20 million generations were performed, with trees sampled every 1,000 generations. All runs were then combined after a burn-in of 10% using LogCombiner v1.7.2. Tracer v1.5[49] was used to verify adequate mixing of the Markov chains and ensure that each parameter had been appropriately sampled (i.e., effective sampling size >200). The maximum credibility tree was determined using TreeAnnotator v1.7.2 and visualized the phylogenic tree with FigTree v1.3.1[50].
The program BEAST was used to estimate divergence times using the Drosophila-Anopheles divergence using a prior distribution normally distributed around a mean of 260 million years ago (mya) and ranging from 243 to 276 mya as suggested in[51]. For comparison, divergence times were also estimated using a mutation rate of 0.0115 mutations per nucleotide per million years, which was estimated from the divergence times and sequence divergence of several insect mt genomes[52].
Results
Sequence data and assembly
This study focuses on analysis of the mt genome of 14 individual Anopheles mosquitoes from seven species. These include 11 individuals from the AP group in PNG (An. punctulatus s.s (n = 5), An. koliensis (n = 2), An. farauti s.s (n = 1, An. farauti 1), An. hinesorum (n = 1, An. farauti 2), An. farauti 4 (n = 2)) and three samples from the An. dirus complex in Thailand (An. dirus s.s (n = 2, An. dirus species A) and An. cracens (n = 1, An. dirus species B) (Table 1).
This study was initiated by sequencing the whole genome of five individual mosquitoes, generating ~37-150 million paired-end reads and resulting in an average 500 X coverage of mtDNA [see Additional file1 and Additional file2]. Each individual mt genome was then de novo assembled. For nine additional mosquitoes, the mt genome was amplified by overlapping long-range PCR products and all samples were simultaneously sequenced on one lane of an Illumina HiSeq 2000 [see Additional file4]. This pooled mtDNA sequencing strategy generated 392,983,105 million paired-end reads of 100 bp resulting in an average 188,000 X coverage of each individual mt genome. Each genome was de novo assembled separately. For each of the newly sequenced genomes, the genes and gene organization (i.e., orientation and order) were identical to that of previously sequenced Anopheles mt genomes[11, 35–37, 43].
Phylogenetic analysis
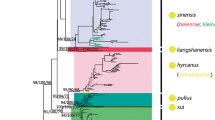
The protein coding DNA sequences of all Anopheles mt genomes generated here, or previously sequenced, were determined and aligned with several outgroups (Table 1). The concatenated protein coding sequence includes 10, 770 nucleotides. A phylogenetic tree was reconstructed using the concatenated protein coding sequences and the Bayesian approach implemented in BEAST[48]. Three independent runs of 20 million iterations were combined and adequate sampling of the posterior distribution of each parameter was obtained. All phylogenetic relationships were supported with posterior probabilities greater than 90%, with the exception of the position of An. gambiae (72% support) and an internal node among the AP mosquitoes (85% support) (Figure 1). The resulting phylogenetic tree highlights three monophyletic clades corresponding to the AP, An. dirus and An. albitarsis groups (Figure 1).
These results show deep divergence between two main Anopheles lineages. One lineage includes all mosquitoes from South and Central America and is further sub-divided into the An. albitarsis complex and An. darlingi species. The other lineage, containing all non-Central and South American Anopheles, seems to have radiated to generate, first Anopheles species currently present in North America and Africa and, from there, SEA and SW Pacific mosquitoes (Figure 2).
Phylogenetic tree of Anopheles using the concatenated DNA sequences of all mitochondrial protein coding genes. The bars illustrate the 95% credibility intervals for the divergence times and the numbers in brackets above each node display the actual values in millions of years. The panel on the right indicates the geographic distribution of the samples: the green bar indicates mosquitoes from South America (SA), red from North America (NA), grey from Africa (AF), blue from Southeast Asian (SEA) and orange from Papua New Guinea (PNG).
The AP group from PNG clusters most closely with the An. dirus complex distributed across SEA (Figure 2). This tree also partially resolves the phylogeny within the AP group with An. farauti 4 being the most divergent while An. farauti s.s. and An. hinesorum being most closely related (Figure 1).
Molecular dating
As a result of the poor fossil record for mosquitoes[53], few reliable calibration points exist for dating anophelines. The divergence times among Anopheles species were therefore estimated using the Drosophila-Anopheles divergence (260 mya[51]). The most recent common ancestor (MRCA) of all Anopheles was dated to 93 mya with a 95% credibility interval ranging from 61 to 126 mya (Table 2). From this origin, Anopheles mosquitoes seem to have rapidly diverged from each other and spread across the globe to reach SEA and the SW Pacific by ~43-87 mya (Table 2).
Within the AP group, a deep divergence among sibling species with the MRCA of the AP group was observed dating back to 25–54 mya, roughly half as old as the MRCA of all Anopheles (Table 2). Importantly, this ancient origin of the AP group does not appear to result from a single highly divergent sibling species: most species analysed here (An. punctulatus s.s., An. koliensis and An. farauti 4) seem to have diverged 25–54 mya, and only An. farauti s.s. and An. hinesorum share a recent common ancestor (5–17 mya) (Figure 2). This finding is especially striking when compared with the only other sequenced Anopheles group: An. albitarsis mosquitoes are distributed across South America (from southern Brazil to Columbia) over a much larger geographic range than the AP group in PNG but share an MRCA dating back to only 6–24 mya, significantly younger than the MRCA of the AP group (Figure 2).
Dating using the estimated insect mt mutation rate of 0.0115 mutations per nucleotide per million years[52] instead of a calibration point, led, overall, to more recent divergence times [see Additional file5] (but see below).
Discussion
The predominant hypothesis regarding the origin of Anopheles mosquitoes predicts that they originated in western Gondwana[10, 11] and that, by 95 mya, Anopheles had migrated into Africa. Ancestral Anopheles are predicted to have then colonized Europe and North America (via land bridges), and migrated through Asia into the SW Pacific. The topology of the tree in Figure 2 is globally consistent with this hypothesis. However, the position of North American mosquitoes (An. quadrimaculatus) relative to African and other non-American Anopheles remains unclear. In particular the lack of mt genomes from European Anopheles mt genomes preclude determining whether African Anopheles are ancestral to European and North-American Anopheles or, alternatively, whether North-American Anopheles derive directly from South-American Anopheles. Additional sampling of mt genomes would provide better resolution of Anopheles early dispersal routes.
Regarding SEA and the SW Pacific, it is generally believed that Anopheles from the SW Pacific derives from SEA mosquitoes[10, 26, 27]. The results of this study suggest that the AP group is most closely related to the An. dirus complex of SEA, consistent with an origin of the AP group in SEA. However, currently it cannot be determined whether SW Pacific and Australian Anopheles originate from an SEA ancestor as currently hypothesized or, alternatively, whether SW Pacific and SEA Anopheles have an Australian origin. The molecular dates in this study, suggest that the ancestor of the AP group was present in PNG between 25 and 54 mya but does not allow rejecting either of these scenarios. Plate tectonic models show that the Australia/PNG plate, that separated from Gondwana during the Cretaceous, moved from a southern position in the Eocene to its current position near SEA[54]. While the upper limit of the age of the AP ancestor (54 mya) corresponds to a time when the distance between PNG and SEA would not have allowed migration between these regions, the lower limit (~25 mya) corresponds to a time when the Australian plate had moved close enough to the Asian plate to enable possible migration of species between the two regions[55]. Inclusion of additional mt genome sequences, in particular from Anopheles complexes restricted to Australia (e.g., Anopheles annulipes) may allow better understanding of these early dispersal routes in SEA and SW Pacific.
The monophyly of AP mosquitoes (Figure 1) suggests that they colonized PNG through a single migration event followed by speciation (as opposed to multiple migrations of pre-established species). This study suggests that the different AP sibling species diverged from each other 25–54 mya, much earlier than proposed in previous studies of the AP group[26, 28]. This deep divergence among AP mosquitoes is unlikely to be caused by a single species that could have diverged from the other sibling species in SEA and colonized PNG later. In fact, most of the AP sibling species are equally divergent from each other, suggesting rapid radiations of AP sibling species upon arrival in the SW Pacific area. Since this analysis relies on a single non-recombining locus, it cannot rule out the possibility that the estimates are influenced by the action of natural selection. However, when only Anopheles mt genomes are analysed, there is little evidence for deviation from a clock-like model of evolution, which suggests that nucleotide substitutions occur at a similar rate on each lineage. Therefore, if natural selection is driving the evolution of the mt genome in Anopheles, it is likely to have acted in a similar manner on all lineages and consequently is unlikely to bias the molecular dates significantly. An additional possible complication is that the phylogenetic tree inferred from mt sequences differs from the actual species tree: since these analyses are based on a single locus, one cannot rule out that incomplete lineage sorting and introgression lead to a phylogenetic reconstruction that does not represent the true evolutionary pathways of the species studied[56, 57]. The long internal branches separating species coupled with the very short branches separating individuals from the same species indicate that incomplete sorting of ancestral polymorphisms is unlikely to affect this phylogeny[58]. Ruling out introgression is difficult without genetic data on multiple unlinked loci (i.e., nuclear), which are complicated to generate due to the lack of a good reference genome sequence (An. gambiae being too divergent), the high divergence among AP species at the nuclear level and the high level of genetic diversity within species. However, there is little evidence of gene flow among AP mosquitoes (but see also Ambrose[28]) and early forced mating studies suggested that most hybrids are non-viable or sterile[15]. Overall, these observations suggest that, for this dataset, the mt gene tree likely recapitulates the actual species tree.
Absolute dates estimated from molecular data should be consider cautiously, especially since these estimates rely on a single calibration point. Note however that the divergence estimates between An. quadrimaculatus and non-American Anopheles, as well as, between An. farauti s.s. and An. hinesorum, are similar to those of previous studies[11, 28, 37]. When the estimated mutation rate for insect mtDNA was used[52], the divergence dates obtained were significantly younger than those obtained using a calibration point [see Additional file5]. This mutation rate, while widely used (see e.g.[28]) was originally calculated using closely related species (the maximum divergence time used in the study was ~3 mya) and, Brower noted, probably overestimated the actual mutation rate. A slower mutation rate would push estimated divergence dates back in time, closer to the estimates obtained using calibration points. Note that even considering these younger divergence dates, the MRCA of AP mosquitoes would still considerably predate the arrival of humans in PNG (see below). Irrespective of the absolute dates, it is interesting to note that the AP group is significantly older than the only other Anopheles group for which several mt genome sequences are available, the An. albitarsis complex (Figure 3). Indeed, the MRCA of the AP group is estimated to be approximately four times older (25–54 mya) than the An. albitarsis complex MRCA (6–24 mya) and almost half as old as the MRCA of all Anopheles mosquitoes. Based on these findings, it can be speculated that after arriving in PNG 25 to 54 mya, the ancestors of AP group became isolated on different islands as sea levels fluctuated and their distribution only overlapped later, when the sea level decreased to merge different islands into current-day PNG.
Given the old divergence times among most AP sibling species, one would expect that most AP species today are reproductively isolated and that hybridization is unlikely to occur in nature, with the possible exception of An. farauti s.s and An. hinesorum that only diverged 5 to 17 mya (see also[28]). This potential reproductive isolation among AP sibling species is supported by early cross-mating experiments suggesting that F1 hybrids between any combination of An. farauti s.s, An. hinesorum, An. koliensis and An. punctulatus s.s are non-viable or sterile in laboratory[15]. These results have important implications for vector control in the SW Pacific. For example, control of malaria by releasing genetically modified-, sterile-, male mosquitoes as was recently proposed[59] would require, in PNG, independent engineering of mosquitoes from, at least, five highly divergent species to significantly impact the populations of the main malaria vectors. In addition, if AP sibling species are reproductively isolated from each other, insecticide resistance arising in one species is unlikely to spread quickly across all AP mosquitoes, but instead resistance mutations would have to occur independently in each species[29]. Further investigations are required to definitively rule out the existence of gene flow among most AP sibling species, as well as to confirm the observation of putative mt genome introgression between An. farauti s.s and An. hinesorum recently described in Southern New Guinea[28].
The ancient divergences among Anopheles species, including among AP from SW Pacific, also raises questions for the evolution of traits related to human malaria transmission. Transmission of plasmodium parasites to humans is facilitated by the anthropophilic behaviour of some Anopheles species. Such behaviour includes usage of human-made habitats as breeding sites and preferential feeding on human blood. Some Anopheles mosquitoes, including most populations of An. gambiae, are highly specialized feeders and rely preferentially on humans for their blood meal[60]. Others are considered more “opportunistic” or “generalist” and the source of their blood meal varies according to several factors including host density (for review see[61]). There are limited data on the feeding preference and behaviour of AP mosquitoes. Early studies of AP mosquitoes seem to indicate a relatively unspecialized feeding behaviour[62, 63].
On the other hand, recent studies by Cooper and colleagues have shown that at least two populations of An. hinesorum show strong feeding preferences and specifically avoid biting humans[22, 28]. Frequency of utilization of human-made habitats for breeding also varies among the AP sibling species[22]. The phylogenetic study described here suggests that the AP sibling species diverged from each other long before humans arrived in PNG, probably ~50,000 years ago[64]. In fact, the molecular dates place the divergence time of AP mosquitoes before the emergence of the Hominidae (or great ape) family[65, 66]. This indicates that any adaptations to humans (for blood meal or larval habitats) would have occurred independently in each AP sibling species, rather than being inherited from a common ancestor, and suggests that co-occurrences of malaria-related traits in Anopheles are the results of convergent evolution[67, 68].
Conclusions
The ancient divergence among AP sibling species, coupled with the recent arrival of humans in PNG, indicates that AP mosquitoes were present in PNG long before humans colonized the island. This observation suggests that the AP mosquitoes have independently evolved to adapt their behaviour to humans. While further studies are needed to better characterize the behaviour of AP mosquitoes, these findings emphasize the potential of the AP group to serve as a model for studying the evolution of vector competence and potentially for identifying the genetic basis of the ability to transmit human malaria.
References
Chaves LF, Harrington LC, Keogh CL, Nguyen AM, Kitron UD: Blood feeding patterns of mosquitoes: random or structured?. Front Zool. 2010, 7: 3-10.1186/1742-9994-7-3.
Hay SI, Sinka ME, Okara RM, Kabaria CW, Mbithi PM, Tago CC, Benz D, Gething PW, Howes RE, Patil AP, Temperley WH, Bangs MJ, Chareonviriyaphap T, Elyazar IR, Harbach RE, Hemingway J, Manguin S, Mbogo CM, Rubio-Palis Y, Godfray HC: Developing global maps of the dominant anopheles vectors of human malaria. PLoS Med. 2010, 7: e1000209-10.1371/journal.pmed.1000209.
Bryan JH: Morphological studies on Anopheles punctulatus Donitz complex. Trans R Entomol Soc London. 1974, 125: 413-435.
Scott JA, Brogdon WG, Collins FH: Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. AmJTrop Med Hyg. 1993, 49: 520-529.
Wilkerson RC, Parsons TJ, Klein TA, Gaffigan TV, Bergo E, Consolim J: Diagnosis by random amplified polymorphic DNA polymerase chain reaction of four cryptic species related to Anopheles (Nyssorhynchus) albitarsis (Diptera: Culicidae) from Paraguay, Argentina, and Brazil. J Med Entomol. 1995, 32: 697-704.
Proft J, Maier WA, Kampen H: Identification of six sibling species of the Anopheles maculipennis complex (Diptera: Culicidae) by a polymerase chain reaction assay. Parasitol Res. 1999, 85: 837-843. 10.1007/s004360050642.
Favia G, della Torre A, Bagayoko M, Lanfrancotti A, Sagnon N, Toure YT, Coluzzi M: Molecular identification of sympatric chromosomal forms of Anopheles gambiae and further evidence of their reproductive isolation. Insect Mol Biol. 1997, 6: 377-383. 10.1046/j.1365-2583.1997.00189.x.
della Torre A, Fanello C, Akogbeto M, Dossou-yovo J, Favia G, Petrarca V, Coluzzi M: Molecular evidence of incipient speciation within Anopheles gambiae s.s. in West Africa. Insect Mol Biol. 2001, 10: 9-18. 10.1046/j.1365-2583.2001.00235.x.
Guelbeogo WM, Grushko O, Boccolini D, Ouedraogo PA, Besansky NJ, Sagnon NF, Costantini C: Chromosomal evidence of incipient speciation in the Afrotropical malaria mosquito Anopheles funestus. Med Vet Entomol. 2005, 19: 458-469. 10.1111/j.1365-2915.2005.00595.x.
Krzywinski J, Besansky NJ: Molecular systematics of Anopheles: from subgenera to subpopulations. Annu Rev Entomol. 2003, 48: 111-139. 10.1146/annurev.ento.48.091801.112647.
Krzywinski J, Grushko OG, Besansky NJ: Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Mol Phylogenet Evol. 2006, 39: 417-423. 10.1016/j.ympev.2006.01.006.
Rozeboom LE, Knight KL: The punctulatus complex of Anopheles (Diptera: Culicidae). J Parasitol. 1946, 32: 95-131. 10.2307/3272584.
Burkot TR, Graves PM, Cattan JA, Wirtz RA, Gibson FD: The efficiency of sporozoite transmission in the human malarias, Plasmodium falciparum and P. vivax. Bull World Health Organ. 1987, 65: 375-380.
Bryan JH: Studies on the Anopheles punctulatus complex. I. Identification by proboscis morphological criteria and by cross-mating experiments. Trans R Soc Trop Med Hyg. 1973, 67: 64-69. 10.1016/0035-9203(73)90322-2.
Bryan JH: Studies on the Anopheles punctulatus complex. II. Hybridization of the member species. Trans R Soc Trop Med Hyg. 1973, 67: 70-84. 10.1016/0035-9203(73)90323-4.
Bryan JH: Studies on the Anopheles punctulatus complex. 3. Mating behaviour of the F1 hybrid adults from crosses between Anopheles farauti no.1 and Anopheles farauti no. 2. Trans R Soc Trop Med Hyg. 1973, 67: 85-91. 10.1016/0035-9203(73)90324-6.
Foley DH, Paru R, Dagoro H, Bryan JH: Allozyme analysis reveals six species within the Anopheles punctulatus complex of mosquitoes in Papua New Guinea. Med Vet Entomol. 1993, 7: 37-48. 10.1111/j.1365-2915.1993.tb00649.x.
Beebe NW, Foley DH, Saul A, Cooper L, Bryan JH, Burkot TR: DNA probes for identifying the members of the Anopheles punctulatus complex in Papua New Guinea. AmJTrop Med Hyg. 1994, 50: 229-234.
Beebe NW, Ellis JT, Cooper RD, Saul A: DNA sequence analysis of the ribosomal DNA ITS2 region for the Anopheles punctulatus group of mosquitoes. Insect Mol Biol. 1999, 8: 381-390. 10.1046/j.1365-2583.1999.83127.x.
Foley DH, Cooper RD, Bryan JH: A new species within the Anopheles punctulatus complex in Western Province, Papua New Guinea. J Am Mosq Control Assoc. 1995, 11: 122-127.
Beebe NW, Saul A: Discrimination of all members of the Anopheles punctulatus complex by polymerase chain reaction–restriction fragment length polymorphism analysis. AmJTrop Med Hyg. 1995, 53: 478-481.
Cooper RD, Waterson DG, Frances SP, Beebe NW, Sweeney AW: Speciation and distribution of the members of the Anopheles punctulatus (Diptera: Culicidae) group in Papua New Guinea. J Med Entomol. 2002, 39: 16-27. 10.1603/0022-2585-39.1.16.
Benet A, Mai A, Bockarie F, Lagog M, Zimmerman P, Alpers MP, Reeder JC, Bockarie MJ: Polymerase chain reaction diagnosis and the changing pattern of vector ecology and malaria transmission dynamics in Papua New Guinea. AmJTrop Med Hyg. 2004, 71: 277-284.
Cooper RD, Waterson DG, Frances SP, Beebe NW, Pluess B, Sweeney AW: Malaria vectors of Papua New Guinea. Int J Parasitol. 2009, 39: 1495-1501. 10.1016/j.ijpara.2009.05.009.
Cooper RD, Waterson DG, Bangs MJ, Beebe NW: Rediscovery of Anopheles (Cellia) clowi (Diptera: Culicidae), a rarely recorded member of the Anopheles punctulatus group. J Med Entomol. 2000, 37: 840-845. 10.1603/0022-2585-37.6.840.
Beebe NW, Cooper RD, Morrison DA, Ellis JT: A phylogenetic study of the Anopheles punctulatus group of malaria vectors comparing rDNA sequence alignments derived from the mitochondrial and nuclear small ribosomal subunits. Mol Phylogenet Evol. 2000, 17: 430-436. 10.1006/mpev.2000.0853.
Foley DH, Bryan JH, Yeates D, Saul A: Evolution and systematics of Anopheles: insights from a molecular phylogeny of Australasian mosquitoes. Mol Phylogenet Evol. 1998, 9: 262-275. 10.1006/mpev.1997.0457.
Ambrose L, Riginos C, Cooper RD, Leow KS, Ong W, Beebe NW: Population structure, mitochondrial polyphyly and the repeated loss of human biting ability in anopheline mosquitoes from the southwest Pacific. Mol Ecol. 2012, 21: 4327-4343. 10.1111/j.1365-294X.2012.05690.x.
Henry-Halldin CN, Nadesakumaran K, Keven JB, Zimmerman AM, Siba P, Mueller I, Hetzel MW, Kazura JW, Thomsen E, Reimer LJ, Zimmerman PA: Multiplex assay for species identification and monitoring of insecticide resistance in Anopheles punctulatus group populations of Papua New Guinea. AmJTrop Med Hyg. 2012, 86: 140-151. 10.4269/ajtmh.2012.11-0503.
Beebe NW, Cooper RD: Distribution and evolution of the Anopheles punctulatus group (Diptera: Culicidae) in Australia and Papua New Guinea. Int J Parasitol. 2002, 32: 563-574. 10.1016/S0020-7519(01)00359-9.
Henry-Halldin CN, Reimer L, Thomsen E, Koimbu G, Zimmerman A, Keven JB, Dagoro H, Hetzel MW, Mueller I, Siba P, Zimmerman PA: High throughput multiplex assay for species identification of Papua New Guinea malaria vectors: members of the Anopheles punctulatus (Diptera: Culicidae) species group. AmJTrop Med Hyg. 2011, 84: 166-173. 10.4269/ajtmh.2011.10-0438.
Langmead B, Trapnell C, Pop M, Salzberg SL: Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10: R25-10.1186/gb-2009-10-3-r25.
Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I: ABySS: a parallel assembler for short read sequence data. Genome Res. 2009, 19: 1117-1123. 10.1101/gr.089532.108.
Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL: Versatile and open software for comparing large genomes. Genome Biol. 2004, 5: R12-10.1186/gb-2004-5-2-r12.
Beard CB, Hamm DM, Collins FH: The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Mol Biol. 1993, 2: 103-124. 10.1111/j.1365-2583.1993.tb00131.x.
Cockburn AF, Mitchell SE, Seawright JA: Cloning of the mitochondrial genome of Anopheles quadrimaculatus. Arch Insect Biochem Physiol. 1990, 14: 31-36. 10.1002/arch.940140104.
Moreno M, Marinotti O, Krzywinski J, Tadei WP, James AA, Achee NL, Conn JE: Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar J. 2010, 9: 127-10.1186/1475-2875-9-127.
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD: Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31: 3497-3500. 10.1093/nar/gkg500.
Rozen S, Skaletsky H: Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000, 132: 365-386.
DNASTAR software for life scientists.http://www.dnastar.com,
Li H, Durbin R: Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009, 25: 1754-1760. 10.1093/bioinformatics/btp324.
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R: The sequence alignment/map format and SAMtools. Bioinformatics. 2009, 25: 2078-2079. 10.1093/bioinformatics/btp352.
Krzywinski J, Li C, Morris M, Conn JE, Lima JB, Povoa MM, Wilkerson RC: Analysis of the evolutionary forces shaping mitochondrial genomes of a neotropical malaria vector complex. Mol Phylogenet Evol. 2011, 58: 469-477. 10.1016/j.ympev.2011.01.003.
Lewis DL, Farr CL, Kaguni LS: Drosophila melanogaster mitochondrial DNA: completion of the nucleotide sequence and evolutionary comparisons. Insect Mol Biol. 1995, 4: 263-278. 10.1111/j.1365-2583.1995.tb00032.x.
Clary DO, Wolstenholme DR: The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. J Mol Evol. 1985, 22: 252-271. 10.1007/BF02099755.
Abascal F, Zardoya R, Telford MJ: TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010, 38: W7-W13. 10.1093/nar/gkq291.
Posada D: jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008, 25: 1253-1256. 10.1093/molbev/msn083.
Drummond AJ, Rambaut A: BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007, 7: 214-10.1186/1471-2148-7-214.
Gaunt MW, Miles MA: An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol Biol Evol. 2002, 19: 748-761. 10.1093/oxfordjournals.molbev.a004133.
Brower AV: Rapid morphological radiation and convergence among races of the butterfly Heliconius erato inferred from patterns of mitochondrial DNA evolution. Proc Natl Acad Sci U S A. 1994, 91: 6491-6495. 10.1073/pnas.91.14.6491.
Poinar GO, Zavortink TJ, Pike T, Johnston PA: Paleoculicis minutus ( Diptera: Culicidae ) n. gen., n. sp., from Cretaceous Canadian amber, with a summary of described fossil mosquitoes. Acta Geologica Hispanica. 2000, 35: 119-128.
Scotese CR: A continental drift flipbook. J Geol. 2004, 112: 729-741. 10.1086/424867.
Hall R: Cenozoic geological and plate tectonic evolution of SE Asia and the SW Pacific: computer-based reconstructions, model and animations. J Asian Earth Sci. 2002, 20: 353-431. 10.1016/S1367-9120(01)00069-4.
Pamilo P, Nei M: Relationships between gene trees and species trees. Mol Biol Evol. 1988, 5: 568-583.
Nichols R: Gene trees and species trees are not the same. Trends Ecol Evol. 2001, 16: 358-364. 10.1016/S0169-5347(01)02203-0.
Degnan JH, Rosenberg NA: Discordance of species trees with their most likely gene trees. PLoS Genet. 2006, 2: e68-10.1371/journal.pgen.0020068.
Harris AF, Nimmo D, McKemey AR, Kelly N, Scaife S, Donnelly CA, Beech C, Petrie WD, Alphey L: Field performance of engineered male mosquitoes. Nat Biotechnol. 2011, 29: 1034-1037. 10.1038/nbt.2019.
Killeen GF, McKenzie FE, Foy BD, Bogh C, Beier JC: The availability of potential hosts as a determinant of feeding behaviours and malaria transmission by African mosquito populations. Trans R Soc Trop Med Hyg. 2001, 95: 469-476. 10.1016/S0035-9203(01)90005-7.
Lyimo IN, Ferguson HM: Ecological and evolutionary determinants of host species choice in mosquito vectors. Trends Parasitol. 2009, 25: 189-196. 10.1016/j.pt.2009.01.005.
Charlwood JD, Dagoro H, Paru R: Blood-feeding and resting behavior in the Anopheles punctulatus Donitz complex (Diptera, Cullicidae) from coastal Papua New Guinea. Bull Entomol Res. 1985, 75: 463-475. 10.1017/S0007485300014577.
Burkot TR, Graves PM, Paru R, Lagog M: Mixed blood feeding by the malaria vectors in the Anopheles punctulatus complex (Diptera: Culicidae). J Med Entomol. 1988, 25: 205-213.
Lilley I: Papua New Guinea’s human past: the evidence of archaeology. Human Biology In Papua New Guinea: The Small Cosmos. Edited by: Attenborough R, Alpers MP. 1992, Oxford: Oxford University Press, 150-171.
Locke DP, Hillier LW, Warren WC, Worley KC, Nazareth LV, Muzny DM, Yang SP, Wang Z, Chinwalla AT, Minx P, Mitreva M, Cook L, Delehaunty KD, Fronick C, Schmidt H, Fulton LA, Fulton RS, Nelson JO, Magrini V, Pohl C, Graves TA, Markovic C, Cree A, Dinh HH, Hume J, Kovar CL, Fowler GR, Lunter G, Meader S, Heger A: Comparative and demographic analysis of orang-utan genomes. Nature. 2011, 469: 529-533. 10.1038/nature09687.
Langergraber KE, Prufer K, Rowney C, Boesch C, Crockford C, Fawcett K, Inoue E, Inoue-Muruyama M, Mitani JC, Muller MN, Robbins MM, Schubert G, Stoinski TS, Viola B, Watts D, Wittig RM, Wrangham RW, Zuberbuhler K, Paabo S, Vigilant L: Generation times in wild chimpanzees and gorillas suggest earlier divergence times in great ape and human evolution. Proc Natl Acad Sci U S A. 2012, 109: 15716-15721. 10.1073/pnas.1211740109.
Besansky NJ, Hill CA, Costantini C: No accounting for taste: host preference in malaria vectors. Trends Parasitol. 2004, 20: 249-251. 10.1016/j.pt.2004.03.007.
White BJ, Collins FH, Besansky NJ: Evolution of Anopheles gambiae in relation to humans and malaria. Annu Rev Ecol Evol S. 2011, 42: 111-132. 10.1146/annurev-ecolsys-102710-145028.
Acknowledgements
We thank the staff of the Entomology Unit of the Papua New Guinea Institute of Medical Research (PNGIMR) and of the Faculty of Tropical Medicine, Mahidol University (Thailand) for their help in collecting Anopheles samples. We also thank the sequencing group of the McGill University and Genome Quebec Innovation Center for their help with Illumina sequencing. This work was supported by Cleveland Clinic funds to DS and the Fogarty International Center (TW007872) and NIAID International Centers of Excellence in Malaria Research Program (5U19AI089686) to PAZ.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contribution
PAZ and DS designed the research, KL, EC and TP performed the experiments and analysed the data, LR, CHH, STS, JS, PMS, PAZ and DS contributed new reagents or analytical tools, KL, EC, STS, PZ and DS wrote the paper with insights from all other co-authors. All authors read and approve the final manuscript.
Electronic supplementary material
12936_2012_2657_MOESM1_ESM.doc
Additional file 1: Sample sequencing information. List of samples sequenced in this study, their sequencing method, read length and the number of paired-end reads generated. (DOC 36 KB)
12936_2012_2657_MOESM2_ESM.pdf
Additional file 2: Coverage of whole genome sequencing reads on mitochondrial and nuclear genes. Coverage per base pair of whole genome sequencing read pairs mapped to 2 mitochondrial genes (COX2 and COI) and one nuclear gene (VGSC). Each color represents one of the mapped read pairs. The numbers in the center of each bar represent the actual coverage per base pair. (PDF 59 KB)
12936_2012_2657_MOESM3_ESM.doc
Additional file 3: Primers used in study. Primers used to amplify mitochondrial genomes by long range PCR, to fill-in gaps between assembled contigs from whole genome sequencing of An. punctulatus and An. farauti 1 (51 bp reads), and to amplify the control region (A + T rich region) of several mitochondrial genomes. Nucleotides in red indicate variable sites among Anopheles. (DOC 50 KB)
12936_2012_2657_MOESM4_ESM.pdf
Additional file 4: Multiplex sequencing method. Diagram of the steps used to amplify and sequence multiple mitochondrial genomes simultaneously on one lane of an Illumina Hiseq 2000 instrument after amplification by long range PCR. (PDF 36 KB)
12936_2012_2657_MOESM5_ESM.doc
Additional file 5: Divergence times using the insect mitochondrial DNA mutation rate. Mean divergence times and 95% credibility intervals for selected nodes using insect mitochondrial DNA mutation rate. (DOC 31 KB)
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Logue, K., Chan, E.R., Phipps, T. et al. Mitochondrial genome sequences reveal deep divergences among Anopheles punctulatus sibling species in Papua New Guinea. Malar J 12, 64 (2013). https://doi.org/10.1186/1475-2875-12-64
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-2875-12-64