Abstract
Porcine reproductive and respiratory syndrome virus (PRRSV) infection has caused huge economic losses in global swine industry over the last 37 years. PRRSV commercial vaccines are not effective against all epidemic PRRSV strains. In this study we performed a high-throughput screening (HTS) of an FDA-approved drug library, which contained 2339 compounds, and found vidofludimus (Vi) could significantly inhibits PRRSV replication in Marc-145 cells and primary porcine alveolar macrophages (PAMs). Compounds target prediction, molecular docking analysis, and target protein interference assay showed that Vi interacts with dihydroorotate dehydrogenase (DHODH), a rate-limiting enzyme in the de novo pyrimidine synthesis pathway. Furthermore, PRRSV infection was restored in the presence of excess uridine and cytidine which promote pyrimidine salvage, or excess orotate which is the product of DHODH in the de novo pyrimidine biosynthesis pathway, thus confirming that the antiviral effect of Vi against PRRSV relies on the inhibition of DHODH. In addition, Vi also has antiviral activity against Seneca virus A (SVA), encephalomyocarditis virus (EMCV), porcine epidemic diarrhea virus (PEDV), and pseudorabies virus (PRV) in vitro. These findings should be helpful for developing a novel prophylactic and therapeutic strategy against PRRSV and other swine viral infections.
Similar content being viewed by others
Introduction
Porcine reproductive and respiratory syndrome (PRRS), also known as porcine blue ear disease, is a highly infectious swine disease worldwide caused by porcine reproductive and respiratory syndrome virus (PRRSV) and characterized by reproductive failure in sows and respiratory diseases in all pigs [1,2,3]. PRRSV infections could be divided into subclinical, lethal, and persistent infections in terms of the pig growth stage and immune status, the virus strains and secondary or co-infected pathogens, environmental conditions, and disease management level [4, 5]. PRRSV is a small enveloped, single-stranded positive-sense RNA virus belonging to the genus Porartevirus, family Arteriviridae, and order Nidovirales [6]. Currently, PRRSV can be classified into two genotypes, i.e., the European genotype and the North American genotype [7]. High genetic diversity is a significant characteristic of PRRSV. PRRSV mutates rapidly at an estimated rate of 3.29 × 10–3 substitutions per nucleotide site per year, developing growing evolutionary strains [8,9,10,11]. The PRRSV strains prevalent in China are mainly the North American genotype.
Nowadays, vaccination is the most common strategy for PRRS prevention and control. Different kinds of vaccines are now in market, such as inactivated vaccine [12], modified-live virus (MLV) vaccine [13], recombinant vector vaccine [14], DNA vaccine [15], and subunit vaccine [16]. However, these commercially available vaccines can provide only incomplete protections [17,18,19]. Therefore, more effective and safe methods for PRRSV control are urgently in need.
Antiviral drugs occupy an important role in disease prevention and control. Natural compounds and compositions provide valuable sources for antiviral drugs, many of which have shown anti-PRRSV activity in vitro, such as proanthocyanidin A2 [20], griffithsin [21], (-)-epigallocatechin-3-gallate [22], tea polyphenols [23], and Emodin [24]. However, so far there is still no commercial antivirals against PRRSV.
In this study, we screened a library of 2339 FDA-approved drugs and firstly identified four new-hit compounds as potential antivirals for PRRSV treatment after three rounds of screening. Among them, vidofludimus significantly inhibited PRRSV infection with the highest select index (SI) by directly targeting dihydroorotate dehydrogenase (DHODH), and showed a general antiviral activity against other swine viruses, demonstrating excellent potential as a broad-spectrum antiviral product.
Materials and methods
Cells, viruses, and reagents
Marc-145 cells (an African green embryonic kidney epithelial cell line, ATCC) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, USA) supplemented with 10% fetal bovine serum (FBS; Gibco, USA) and Penicillin (250 U/mL)-Streptomycin (250 μg/mL) at 37 °C in a humidified atmosphere containing 5% CO2. Porcine alveolar macrophages (PAMs) were collected from lung lavages of 6-week-old Yorkshire pigs (free of PRRSV, PCV2, PRV), as previously described [11], and cultured in Roswell Park Memorial Institute 1640 medium (RPMI 1640; Gibco, USA) containing 10% FBS and Penicillin (250 U/mL)-Streptomycin (250 μg/mL) at 37 °C in a humidified atmosphere containing 5% CO2. Three North American genotype PRRSV strains were employed. The highly pathogenic PRRSV strain BB0907 (GenBank accession no. HQ315835.1) was used for all experiments and represented as “PRRSV” in this article unless otherwise specified. The PRRSV strains S1 (a classical strain; GenBank accession no. DQ459471.1) and FJ1402 (a NADC30-like strain; GenBank accession no. KX169191.1) were used and named as S1 and FJ1402, respectively. These PRRSV strains were all maintained in our laboratory. PEDV YZ (GenBank accession no. MK841495.1), PRV ZJ01 (GenBank accession no. KM061380.1), SVA CH-SD (GenBank accession no. MH779611.1), and EMCV NJ08 (GenBank accession no. HM641897) were maintained in laboratory. Vidofludimus (Vi), dihydroorotate (DHO), orotate (ORO), uridine, and cytidine were purchased from Selleck Chemicals (purity > 99%; Selleck Chemicals, USA). 6-azauracil (6-AU), an inhibitor of orotidine 5'-phosphate decarboxylase (ODCase), was purchased from Sigma (purity > 98%; Sigma-Aldrich, USA).
Screening of a natural product library
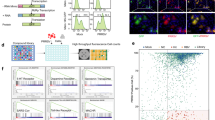
An FDA-approved library containing 2339 compounds was purchased from Selleck Chemicals and stored as 10 mM stock solutions in DMSO at -80 °C until use. The workflow for high-throughput screening (HTS) was carried out as shown in Figures 1A and B. Marc-145 cells were infected with PRRSV (0.01 multiplicity of infection (MOI)) or mock-infected. Drug treatment (5 µM compound or DMSO) was performed from 1 h before virus infection until cells were collected for cytopathic effect (CPE) observation and indirect immunofluorescence assay (IFA) analysis at 48 h post-infection (hpi). Fluorescence intensity was measured by ImageJ software. The inhibition rate of each compound was normalized to the equal volume of DMSO control group. Each assay was performed in duplicate.
High-throughput screening (HTS) for inhibitors of PRRSV infection from an FDA-approved drug library. A HTS assay timeline. B HTS assay flowchart. C Each dot represents the percent inhibition of PRRSV (0.01 MOI) achieved with by compound (5 μM). D IFA of infected Marc-145 cells treated with one of the four designated compounds. PRRSV N protein is colored green, and brightfield-imaged cells show CPE. Scale bars, 500 μm. E CC50 and EC50 of Ta (i), Vi (ii), Be (iii) and Co (iv). Insets show the structure of each compound.
During the primary screening, compounds were weed out if they showed any observable cytotoxicity or demonstrated a less than 50% reduction of CPE compared to the DMSO control group. For the second round of screening, compounds displaying an over 80% of cell viability and the inhibition rate of PRRSV infection were selected. In the final round of screening, 50% effective concentration (EC50) and 50% cytotoxic concentration (CC50) of each remaining candidate compound were calculated using the log (inhibitor) vs. response—Variable slope (four parameters) method by GraphPad Prism 7.0 software (GraphPad Software; CA, USA). The compounds which displayed a dose-dependent inhibition activity on PRRSV infectivity and a selectivity index (SI) higher than ten were considered for further study.
Cell viability assay
Marc-145 cells were treated with compounds or transfected with siRNAs, and incubated for 48 h at 37 °C in a humidified atmosphere containing 5% CO2. Cell viability was tested using an enhanced Cell Counting Kit-8 (CCK-8; Beyotime, China) following the manufacturer’s instructions. The CC50 was calculated using GraphPad Prism 7.0 software. DMSO was used as the negative control.
PRRSV infectivity inhibition assay
Indirect immunofluorescence assay (IFA) was used to examine the effect of the compounds, including tamoxifen citrate (Ta), vidofludimus (Vi), betulonic acid (Be), and corylin (Co), on PRRSV infectivity. Cells were treated with two-fold serially diluted compounds (1 µM to 10 µM), and infected with PRRSV (0.01 MOI) for 48 h at 37 °C. Cells were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.1% Triton X-100 for 30 min at 37 ℃. PRRSV infectivity was detected with a mouse anti-PRRSV N-protein mAb (1:200 dilution, made in laboratory [25]) as primary antibody and the Alexa Fluor 488-conjugated goat anti-mouse IgG (H–L) (1:200 dilution; Proteintech, USA) as secondary antibody. Nuclei were stained with DAPI (Beyotime, China) for 10 min at room temperature. Immunofluorescence was observed using a Zeiss inverted fluorescence microscope. Fluorescence intensity was determined by ImageJ software. The EC50 of each compound was estimated by GraphPad Prism 7.0 software. The SI was determined by the ratio of CC50 to EC50.
Western blot assay
Cells were lysed with 100 μL of Radio-immunoprecipitation assay (RIPA) lysis buffer (Beyotime, China) for 15 min on ice, then resolved by SDS-PAGE and transferred to a nitrocellulose membrane. The membrane was then blocked with 5% non-fat milk in PBST (w/v) and then probed with the following antibodies: anti-PRRSV N-protein mAb (1:1000 dilution), anti-SVA VP2-protein mAb (1:1000 dilution), anti-EMCV VP1-protein mAb (1:1000 dilution), anti-PEDV N-protein mAb (1:1000 dilution), or anti-PRV gB-protein mAb (1:1000 dilution) prepared in our laboratory; and anti-β-actin mAb (1:10 000 dilution; Proteintech, USA), anti-DHODH mAb (1:2000 dilution; Proteintech, USA), or anti-HA mAb (1:5000 dilution; BioWorld, USA) as primary antibody, respectively. Horseradish peroxidase (HRP)-conjugated goat anti-rabbit or goat anti-mouse IgG (H–L) were used as secondary antibodies (1:1000 dilution; Beyotime, China).
RNA extraction and quantitative real‑time PCR
Total RNA was extracted from cells using a Total RNA Kit I (Omega Bio-Tek, USA). RNA was then reverse transcribed into cDNA using HiScript qRT SuperMix (Vazyme, China) following the manufacturer’s instructions. Quantitative RT-PCR was performed using an ABI QuantStudio 6 Systems (Applied Biosystems, USA) with AceQ® qPCR SYBR® Green Master Mix (Vazyme, China) following the manufacturer’s instructions. Gene quantification was referenced to monkey GAPDH or pig β-actin genes, normalized to the mock-infected control and calculated through 2−ΔΔCt methods and the results were calculated as mean ± standard deviation (SD). The primers are listed in Table 1.
Virus titration
Cells were infected with ten-fold serial dilutions of indicated viruses in eight replicates. After 1 h inoculation at 37 °C, the cells were washed and incubated with fresh medium for 2–5 days. Viral titers were determined using endpoint dilution analysis five days post-inoculation (dpi). The cytopathic effect was observed using an inverted microscope. The median tissue culture infectious dose (TCID50) was determined by the Reed-Muench method.
Virucidal activity assay
To evaluate the virucidal activity of Vi, Vi (10 µM) or DMSO was incubated with PRRSV (0.1 and 1 MOI) for 3 h at 37 °C. The mixtures were then subjected to virus titration as described above.
Virus binding assay
Marc-145 cells or PAMs were pre-chilled for 1 h at 4 °C before treated with Vi (5 or 10 µM) for 1 h at 4 °C. Then the cells were infected with the mixture of PRRSV (1 MOI) and Vi (5 or 10 µM) for 1 h at 4 °C. Cells were washed 3 times with ice-cold PBS before viral RNA were extracted and quantified by qRT-PCR as described above.
Virus internalization assay
Marc-145 cells pre-treated with cycloheximide (CHX; 10 μg/mL) for 12 h before PRRSV infection (1 MOI) for 1 h at 4 °C to allow virus attachment. Cells were washed 3 times with ice-cold PBS to remove unbound virus, following by 2 h incubation with fresh DMEM containing 10 µM Vi or DMSO at 37 °C. Cells were washed with citrate buffer (pH 3.0) to remove the non-internalized virus and viral RNA were extracted and quantified by qRT-PCR as described above.
Virus replication assay
Marc-145 cells were infected with PRRSV (1 MOI). At 6 hpi, the cells were washed 3 times with PBS and then incubated with fresh medium containing Vi (10 µM) or DMSO at 37 °C. Viral RNA were extracted and quantified by qRT-PCR at indicated time post infection as described above.
Virus release assay
Marc-145 cells were infected with PRRSV (0.1 MOI). At 24 hpi, cells were washed 3 times with PBS and incubated with fresh medium containing Vi (10 µM) or DMSO for 10, 30, and 60 min at 37 °C. Cell supernatants were harvested at indicated time points for virus titration as described.
Effect of DHODH overexpression on PRRSV replication
DHODH gene was amplified with cDNA from Marc-145 cells and cloned into the pCAGGS with an HA tag at its 3’ end to produce pCAGGS-chloDHODH. The primers are listed in Table 1. Marc-145 cells were transfected with 0, 0.1, 0.2, 0.3, or 0.5 μg of pCAGGS-chloDHODH using Lipofectamine™ 3000 (Invitrogen, USA) according to the manufacturer’s recommendations. At 24 h post-transfection (hpt), the cells were infected with PRRSV (0.1 MOI) or treated with Vi (3 μM) and then infected with PRRSV (0.1 MOI). Cells were harvested at 48 hpi for Western blotting detection as described above.
Effect of DHODH interference on PRRSV replication
Marc-145 cells were transfected with 100 nM of siDHODH (Biotend, China) or negative control (siNC) using Lipofectamine™ 3000 reagent (Invitrogen, USA). At 18 hpt, the cells were treated with Vi and infected with PRRSV (0.4 MOI). Cells were harvested at 36 hpi for Western blotting detection as described above. The sequences of siDHODH were shown in Table 1.
Target proteins prediction using SwissTargetPrediction
The structure of Vi was analyzed by the PubChem database and put into the SwissTargetPrediction for identification of potential drug targets in Homo sapiens [26,27,28].
In silico docking
The crystal structure of rat DHODH (ratDHODH) was obtained from the Protein Data Bank (PDB: 1UUO). The putative 3D structure of chlorocebus sabaeus DHODH (chloDHODH) and sus scrofa DHODH (susDHODH) were predicted and scored using the online tool SWISS-MODEL [29]. Quality assessments of the predicted 3D models, including Ramachandran plot score and Z-score, were performed using the online tools SAVES v6.0 and ProSA-web [30,31,32]. The 3D structure of vidofludimus (Vi) was obtained from PubChem (Compound CID: 9820008).
The Autodock 4.2 program (genetic algorithm) was used for the docking of Vi to chloDHODH. The estimated free energy of binding was ranked, and the top one complex was employed. The docking results were visualized using PyMOL 2.3.2.
Molecular dynamic simulation
Thermodynamic constancy of the receptor-ligand system was analyzed through the Gromacs2021.2 software [33, 34]. Firstly, AmberTools22 was used to add GAFF force field to the small molecule. Gaussian 16W carried out hydrogenation of small molecules and calculation of RESP potential, and the potential data would be added to the molecular dynamics system topology file. Simulations were conducted with the Gromacs package using Amber99sb-ildn force field at static temperature 300 K and 1 bar pressure. Long-range electrostatic interactions were treated with the particle-mesh Ewald method. The Tip3p water model was used to solvate the protein in a periodic dodecahedron box extending 10 Å from the nearest protein atom. The total charge of the simulation system was balanced by adding an appropriate amount of Na+, minimized by the steepest descent method, and equilibrated with isothermal isovolumic ensemble (NVT) and isothermal isobaric ensemble (NPT) for 100 000 steps, respectively, with the coupling constant of 0.1 ps and the duration of 100 ps. All bond lengths were constrained with the LINear Constraint Solver algorithm. A cut-off of 14 Å was used to calculate short-range van der Waals and electrostatic interactions. Finally, the free molecular dynamics simulation was performed. The time step was 2 fs and the total simulation time was 100 ns. The root-mean-square deviation (RMSD) and the number of hydrogen bonds between ligand and active pockets of the proteins were analyzed to judge binding stability and convergence.
Broad‑spectrum antiviral assessment
Western blot and TCID50 were performed to examine the Vi antiviral activity against other swine disease viruses. BHK-21 cells were infected with SVA (0.02 MOI) or EMCV (0.02 MOI), Vero cells were infected with PEDV (0.1 MOI), and PK-15 cells were infected with PRV (0.1 MOI), with the addition of Vi (0–10 μM) in culture medium. The cells and supernatants were harvested at different time points as indicated.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software, USA). Results are expressed as the mean ± standard deviation (SD). The significance of differences among groups was determined by one-way or two-way analysis of variance (ANOVA). The asterisks indicate significant differences (*P < 0.05; ** P < 0.01; *** P < 0.001; **** P < 0.0001; ns, not significant).
Results
Library screening
In order to detect the effect of the compounds on PRRSV infection, Marc-145 cells were treated with 5 µM compounds and infected with PRRSV as illustrated in Figure 1A. After primary screening, 61 (2.61%) compounds showing no apparent cytotoxicity and 50% CPE reduction compared to the DMSO group were found. These 61 compounds were then subjected to a second round of screening and 25 compounds leading to negligible cytotoxicity and over 80% inhibition rate were screened. After a final screening with the 25 compounds, 4 compounds, including tamoxifen citrate (Ta), vidofludimus (Vi), betulonic acid (Be) and corylin (Co), showed PRRSV inhibition activity in a dose-dependent manner and exhibited an SI higher than 10 (Figures 1B–E, Table 2). Vi was selected for further study as it showed a highest SI of 24.01.
Vidofludimus inhibits PRRSV infection
The cytotoxicity of Vi was detected on Marc-145 cells with different concentrations of Vi from 1 to 16,000 nM. As shown in Figure 2A, the cell viability of Marc-145 cells was less than 80% until Vi reaches 16 µM. Therefore, the anti-PRRSV activity of Vi were detected with 1, 2.5, 5, and 10 µM. Western blotting of PRRSV N protein, qRT-PCR of ORF7 mRNA, TCID50 analysis, CPE, and IFA observation all showed a dose-dependent antiviral activity of Vi against PRRSV (Figures 2B–E). The anti-PRRSV activity of Vi was next confirmed with PRRSV NADC30-like strain FJ1402 and the classical strain S1. The results showed a general antiviral activity of Vi against different PRRSV strains (Figures 2F, G), and the EC50 of Vi on PRRSV FJ1402 and PRRSV S1 were 3.31 μM and 2.56 μM, respectively (Figures 2H, I).
Vidofludimus anti‑PRRSV activity in Marc‑145 cells. A Viability of Marc-145 cells treated with the indicated concentrations of Vi for 48 h. B Western blot of N-protein in cells infected with PRRSV and treated with the indicated concentrations of Vi. C Relative PRRSV ORF7 mRNA levels determined by qRT-PCR. GAPDH was used as reference control. D Virus titration by TCID50 calculation. E Light microscopy and IFA detection of Marc-145 cells (PRRSV-infected and Vi-treated), at 48 hpi. Green, PRRSV N-protein; blue, nucleus. Scale bars, 500 μm. F and G Western blot of N-protein and TCID50 detection in cells infected with different PRRSV genotypes Vi or DMSO treatment. H EC50 of Vi on PRRSV NADC30-like strain FJ1402. I EC50 of Vi on PRRSV classical strain S1. Error bars represent the mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns, not significant.
The anti-PRRSV activity of Vi was further explored in primary porcine cells, porcine alveolar macrophages (PAMs). Cell viability analysis showed that Vi exhibited little cytotoxicity on PAMs at various concentrations until 16 000 nM (Figure 3A). Western blotting of PRRSV N protein, qRT-PCR of ORF7 mRNA, and TCID50 analysis showed a significant and dose-dependent anti-PRRSV activity of Vi in PAMs (Figures 3B–D).
Vidofludimus anti‑PRRSV activity in PAMs. A Viability of PAMs treated with the indicated concentrations of Vi for 48 h. B Western blot of N-protein in cells infected with PRRSV and treated with the indicated concentrations of Vi. C Relative PRRSV ORF7 mRNA levels determined by qRT-PCR. GAPDH was used as reference control. D Virus titration by TCID50 calculation. Error bars represent the mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns: not significant.
Vidofludimus inhibits PRRSV infection during virus binding and replication stage
Marc-145 cells were infected with PRRSV with Vi treatment at different stages as shown in Figure 4A. The results showed that Vi exhibited no elimination activity against PRRSV in vitro (Figure 4B), and did not affect PRRSV infection during virus internalization and release stage (Figures 4D and F). Interestingly, when Vi was added into the Marc-145 cells during virus binding and replication stage, quantification of PRRSV ORF7 mRNA showed a significantly reduction in the Vi-treatment group compared to the DMSO-treatment group, indicating that Vi inhibited PRRSV infection during virus binding and genome replication stage in Marc-145 cells (Figures 4C, E). Meanwhile, Vi also significantly inhibited PRRSV binding and replication in PAMs cells (Figures 4G, H).
Time-of-addition analysis of vidofludimus anti-PRRSV activity. A Schematic illustration of the time-of-addition experiment. DMSO was used as the solvent of Vi. And an equal volume of DMSO was used for treating viruses as black solvent control. B TCID50 detection for virucidal activity assay. C PRRSV ORF7 mRNA levels determination by qRT-PCR in virus binding assay. GAPDH was used as reference control. D PRRSV ORF7 mRNA levels determination by qRT-PCR in virus internalization assay. GAPDH was used as reference control. E PRRSV ORF7 mRNA levels determination by qRT-PCR in virus replication assay. GAPDH was used as reference control. F Virus titration by TCID50 calculation with cell supernatant in virus release assay. G PRRSV ORF7 mRNA levels determination by qRT-PCR in virus binding assay in PAMs. β-actin was used as reference control. H PRRSV ORF7 mRNA levels determination by qRT-PCR in virus replication assay in PAMs. β-actin was used as reference control. Error bars represent the mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns: not significant.
Vidofludimus stably binds to DHODH in silico
In order to explore the molecular mechanism that Vi inhibited PRRSV infection, the target host proteins of Vi was predicted by SwissTargetPrediction. Ten host proteins were exported as possible targets against Vi, of which the oxidoreductase dihydroorotate dehydrogenase (DHODH) showed a probability of 1 (Table 3). The structure of chlorocebus sabaeus DHODH (chloDHODH) was predicted using the online tool SWISS-MODEL and the reliability of the predicted structure was analyzed on SAVES v6.0 and ProSA-web. The Ramachandran plot analysis of chloDHODH revealed 94.1, 5.9, 0, and 0% residues in the most favorable, additional allowed, generously allowed, and disallowed regions, respectively (Additional file 1A), and a Z-score value of -9.57 (Additional file 1B). The binding activity between Vi and chloDHODH was analyzed using Autodock. Vi was shown to bind chloDHODH with a binding energy of -9.58 kcal/mol (Figure 5A). The binding stability was further analyzed by Gromacs2021.2 software through measurement of RMSD value, of which 0.1–0.3 nm indicated a relatively stable binding of the complex [35], and the hydrogen bonds numbers. The RMSD value of Vi-chloDHODH complex tended to be stable after 23 ns and stayed lower than 0.3 nm during the 100 ns (Figure 5B), and a generally 3–4 hydrogen bonds were formed between Vi and chloDHODH (Figure 5C), indicating a stable interaction between Vi and chloDHODH.
Target analysis of vidofludimus. A Docked conformation of chloDHODH with Vi. The compound and protein are represented as sticks and cartoons, respectively. Vi is colored orange and the protein chloDHODH is colored green. The binding site is shown as cavity structure. The binding energy of the Vi-chloDHODH complex is marked with an asterisk. B RMSD values of chloDHODH (black), Vi (red), and complex (Vi & chloDHODH, blue) over the 100 ns simulation time. C Number of hydrogen bonds involved in the interaction between chloDHODH and Vi during the MD simulation.
Vidofludimus inhibited PRRSV replication through suppression of UMP synthesis in host cells
As DHODH was possibly a host target of Vi, we next investigated the role of DHODH during PRRSV replication. The overexpression of chloDHODH in Marc-145 cells showed a dose-dependent promotion activity on PRRSV replication (Figure 6A), and knockdown of chloDHODH gene by siRNA-1/3 restrained PRRSV replication (Figures 6B, C). When Vi was added into the culture medium, significant reduction of PRRSV N protein levels only occurred in the non-interference and si-NC groups but not in si-DHODH group (Figures 6D–F), indicating that the antiviral activity of Vi was achieved by targeting DHODH, which is consistent with the results of computer simulation analysis. Meanwhile, as shown in Figures 6G, H, Vi treatment reversed the effect of overexpressed DHODH on PRRSV replication. 6-azauracil (6-AU) is a potent inhibitor of Orotidine 5’-monophosphate decarboxylase (ODCase), which serves as a downstream enzyme of DHODH, catalyzing orotidine 5'-monophosphate (OMP) to uridine 5'-monophosphate (UMP). Further investigation showed that 6-AU could also inhibited PRRSV replication in a dose-dependent manner (Figures 6I, J), giving a hint that DHODH affected PRRSV replication through its activity in UMP synthesis.
Vidofludimus anti-PRRSV activity is mediated by DHODH. A Effect of DHODH overexpression on PRRSV replication in Marc-145 cells. B Viability of Marc-145 cells treated with the indicated concentrations of siRNAs for 48 h. C Effect of DHODH interference on PRRSV replication in Marc-145 cells. D Effect of DHODH interference on Vi anti-PRRSV activity in Marc-145 cells. E and F Image J analysis of DHODH and PRRSV N protein quantification in (D). G Effect of Vi treatment on DHODH proviral activity in Marc-145 cells. H Image J analysis of PRRSV N protein quantification in (G). I Viability of Marc-145 cells treated with the indicated concentrations of 6-AU for 48 h. J Western blot of N-protein in PRRSV-infected cells with indicated concentrations of 6-AU or DMSO.
DHODH is an oxidoreductase catalyzing dihydroorotate to orotate for UMP synthesis [36]. To further verify if chloDHODH affects PRRSV replication through its activity in UMP synthesis, a series of host molecules in UMP synthesis pathway were detected for their effect in Vi-PRRSV interaction. As shown in Figure 7, addition of dihydroorotate (DHO) did not reverse the inhibition activity of Vi on PRRSV replication. However, addition of orotate (ORO), uridine, and cytidine broke the anti-PRRSV activity of Vi in a dose-dependent manner. These results suggested that Vi inhibited PRRSV replication through the UMP synthesis pathway by interaction with DHODH.
Effect of DHO, ORO, uridine, and cytidine on vidofludimus anti-PRRSV activity. Marc-145 cells were infected with PRRSV (0.1 MOI) with treatment of Vi and indicated concentrations of DHO, ORO, uridine, or cytidine for 48 h. DMSO served as the treatment control. A, C, E, and G Viability of Marc-145 cells treated with the indicated concentrations of DHO, ORO, uridine, or cytidine. B, D, F, and H Western blot of N-protein in cells infected with PRRSV and treated with the indicated compounds or DMSO.
Vidofludimus has broad-spectrum antiviral activity against other swine disease viruses
The antiviral activity of Vi was further investigated with Seneca valley virus (SVA), encephalomyocarditis virus (EMCV), porcine epidemic diarrhea virus (PEDV) and pseudorabies virus (PRV). As shown in Figure 8, Vi exhibited a dose-dependent antiviral activity against SVA, EMCV, PEDV, and PRV within the safe concentration range. Structural analysis of ratDHODH in BHK-21 cells, chloDHODH in Vero cells, and susDHODH in PK-15 cells showed a highly coincident structure among them (Additional files 2A and B), and the binding between Vi and ratDHODH/susDHODH showed a binding energy of -9.23 and -8.32 kcal/mol, respectively (Additional file 2C), indicating that the antiviral activity of Vi against SVA, EMCV, PEDV and PRV was also associated with the DHODH-mediated pyrimidine metabolism.
Broad-spectrum antiviral activity of vidofludimus against SVA, EMCV, PEDV, and PRV. A Viability of BHK-21, Vero, and PK-15 cells pretreated with the indicated concentrations of Vi for 18 h, 16 h, and 20 h, respectively. Western blot and TCID50 were used to examine the inhibition activity of Vi against SVA (B and C), EMCV (D and E), PEDV F and G and PRV (H and I). Error bars represent the mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns: not significant.
Discussion
PRRSV is one of the most important causative agents in swine production worldwide and causes huge economic losses every year [37]. Unfortunately, the current vaccines and immunization strategies cannot be effectively protective due to the high diversity of PRRSV strains, Therefore, novel strategies with a broad spectrum of protectiveness are urgently in need. In this study, an FDA-approved drug library was screened and four-hit compounds were firstly identified with anti-PRRSV activity from 2339 compounds. Among them, Vi exhibited a most substantial antiviral effect with the highest select index of 24.01.
Vi is a multifunctional molecule with immunoregulation and anti-inflammatory activity, being a potential treatment option for SARS-CoV-2 [38,39,40]. Here the anti-PRRSV activity of Vi was identified and the mechanisms were further explored. Vi showed an antiviral activity against different genotypes of PRRSV with little cytotoxicity to host cells. A time-of-addition analysis showed that Vi inhibited PRRSV infection in virus replication stage. Vi was predicted as a conjugate of DHODH, which is a key rate-limiting enzyme during the de novo synthesis of pyrimidine. Further exploration found that DHODH was an important promoter for PRRSV infection as overexpression of DHODH significantly promoted PRRSV replication while knockdown of DHODH significantly inhibited PRRSV replication. Moreover, knockdown of DHODH eliminated the antiviral activity of Vi against PRRSV, indicating that Vi might suppress PRRSV replication through DHODH. As 6-AU, a potent inhibitor of ODCase, inhibited PRRSV replication in a dose-dependent manner, it was suspected that Vi-DHODH effect on PRRSV replication could also be associated with the UMP synthesis. Addition of a series of host molecules in UMP synthesis pathway during Vi treatment on PRRSV infection confirmed that the anti-PRRSV activity of Vi was accomplished by suppressing UMP synthesis through blocking the oxidoreductase catalyzing activity of DHODH. As UMP synthesis is an important intermediate link to pyrimidine synthesis in mitochondria, it is believed that the host pyrimidine synthesis should be vital for PRRSV replication. As early as 2010, Kulkarni et al. found that 4SC-101 (synonym of Vi) significantly inhibits DHODH enzyme activity in humans, rats, and mice [41]. Subsequently, multiple research teams independently reported that DHODH inhibitor Vi has therapeutic effects on various diseases, including systemic lupus erythematosus, inflammatory bowel disease, renal transplantation rejection reaction, and relapsing–remitting multiple sclerosis [38, 39, 41, 42]. In recent years, there have been reported that DHODH inhibitors IMU-838 (synonym of Vi) and teriflunomide could antagonize viral infections such as SARS-CoV-2 and West Nile virus, respectively [40, 43]. In order to confirm the Vi treatment affecting pyrimidine biosynthesis, we used the E. coli expression system to express chloDHODH for conducting a biochemical experiment. However, we could not obtain the chloDHODH protein with enzymatic activity. This experiment should be done to confirm the Vi treatment affecting pyrimidine biosynthesis in the future.
DHODH is a general antiviral target as some antivirals with broad spectrums against negative-sense RNA viruses, positive-sense RNA viruses, DNA viruses, retroviruses, flaviviruses, cytomegaloviruses, adenoviruses, and coronaviruses have also been reported to play a DHODH-mediated antiviral activity [44,45,46,47]. This gives a hint that the antiviral activity of Vi might also be broad-spectrum. Further research in this study confirmed this as Vi exhibited a dose-dependent antiviral activity against SVA, EMCV, PEDV, and PRV in different cell types originated from various species. We next analyzed the structures of DHODH proteins from different species, and the binding activities between them and Vi. It showed that the DHODH proteins of rat, sus scrofa, and chlorocebus sabaeus showed a highly conserved structure and a strong interaction possibility with Vi (Additional file 2 and Figure 5). These results suggested that Vi could be an effective antiviral against various viruses whose replication rely on the DHODH-mediated host pyrimidine synthesis.
Our results also showed that Vi could inhibit PRRSV infection in virus binding stage. As heparan sulfate proteoglycan 2 (HSPG2), syndecan-4 (Sdc-4), CD163, and heparanase (HPSE) are closely included during PRRSV binding and entry [48,49,50], the mRNA synthesis of these four genes were detected during Vi suppression on PRRSV infection. The results showed that the mRNAs of HSPG2, Sdc-4, and CD163, but not HPSE, were significantly downregulated after Vi treatment (Additional file 3), indicating that HSPG2, Sdc-4, and CD163 might also be the targets of Vi for its antiviral activity. Further mechanisms are still under exploration.
As concluded in Figure 9, Vi, a potent DHODH inhibitor, effectively inhibited PRRSV and other swine viruses’ infection by blocking the de novo pyrimidine biosynthesis pathway. These findings provide a new and promising therapeutic possibility of Vi for combating infections caused by pathogens in need of host pyrimidines. Future research should focus on the assessment and improvement of the antiviral activity of DHODH inhibitors in susceptible hosts as well as their security and efficacies in vivo in combination with vaccines for disease control.
The schematic diagram of PRRSV inhibition by vidofludimus. Dihydroorotate dehydrogenase (DHODH), the key rate-limiting enzyme of the fourth step reaction in the de novo synthesis of pyrimidine (blue arrow), is important in generating UMP required for viral replication. By targeting and inhibiting DHODH activity, vidofludimus (Vi) blocks the production of orotate (ORO) and finally inhibits the synthesis of viral RNA. In addition, the effect of DHODH inhibitors can be disturbed by the salvage pathway (green arrow). On the other hand, Vi can also inhibit PRRSV adsorption on target cells, and its mechanism may be related to PRRSV adsorption-related protein receptors.
Availability of data and materials
The data and materials will be made available on reasonable request.
Abbreviations
- OMP:
-
orotidylic acid
- UMP:
-
uridine monophosphate
- UDP:
-
uridine diphosphate
- UTP:
-
uridine triphosphate
- CTP:
-
cytidine triphosphate
- CDP:
-
cytidine diphosphate
- CMP:
-
cytidine monophosphate
- CAD:
-
complex of the following three enzymes: carbamoyl-phosphate synthetase 2, aspartate carbamoyl transferase and dihydroorotase
- DHO:
-
dihydroorotate
- DHODH:
-
dihydroorotate dehydrogenase
- UMPS:
-
uridine monophosphate synthetase
- ORO:
-
orotate
- QH2 :
-
ubiquinol
- Q:
-
ubiquinone
- ODCase:
-
orotidine 5'-phosphate decarboxylase
- 6-AU:
-
6-azauracil
- CMPK:
-
cytidine/uridine monophosphate kinase
- NDPK:
-
nucleoside diphosphate kinase
- CTPS:
-
cytidine triphosphate synthetase
- UPP 1/2:
-
uracil phosphoribosyltransferase 1 and uracil phosphoribosyltransferase 2
- CDA:
-
cytidine deaminase
- UCK 1/2:
-
uridine-cytidine kinase 1 and uridine-cytidine kinase 2
- UPRT:
-
uracil phosphoribosyl transferase
References
Paton DJ, Brown IH, Edwards S, Wensvoort G (1991) “Blue ear” disease of pigs. Vet Rec 128:617
Pejsak Z, Stadejek T, Markowska-Daniel I (1997) Clinical signs and economic losses caused by porcine reproductive and respiratory syndrome virus in a large breeding farm. Vet Microbiol 55:317–322
Lunney JK, Fang Y, Ladinig A, Chen N, Li Y, Rowland B, Renukaradhya GJ (2016) Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu Rev Anim Biosci 4:129–154
Snijder EJ, Kikkert M, Fang Y (2013) Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163
Albina E (1997) Epidemiology of porcine reproductive and respiratory syndrome (PRRS): an overview. Vet Microbiol 55:309–316
Adams MJ, Lefkowitz EJ, King AMQ, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR, Nibert M, Sabanadzovic S, Sanfacon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Gorbalenya AE, Davison AJ (2017) Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch Virol 162:2505–2538
Kuhn JH, Lauck M, Bailey AL, Shchetinin AM, Vishnevskaya TV, Bao Y, Ng TF, LeBreton M, Schneider BS, Gillis A, Tamoufe U, Diffo Jle D, Takuo JM, Kondov NO, Coffey LL, Wolfe ND, Delwart E, Clawson AN, Postnikova E, Bollinger L, Lackemeyer MG, Radoshitzky SR, Palacios G, Wada J, Shevtsova ZV, Jahrling PB, Lapin BA, Deriabin PG, Dunowska M, Alkhovsky SV, Rogers J, Friedrich TC, O’Connor DH, Goldberg TL (2016) Reorganization and expansion of the nidoviral family Arteriviridae. Arch Virol 161:755–768
Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF (2007) Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526
Li Y, Wang X, Bo K, Wang X, Tang B, Yang B, Jiang W, Jiang P (2007) Emergence of a highly pathogenic porcine reproductive and respiratory syndrome virus in the Mid-Eastern region of China. Vet J 174:577–584
Song J, Shen D, Cui J, Zhao B (2010) Accelerated evolution of PRRSV during recent outbreaks in China. Virus Genes 41:241–245
Zhang Q, Bai J, Hou H, Song Z, Zhao Y, Jiang P (2017) A novel recombinant porcine reproductive and respiratory syndrome virus with significant variation in cell adaption and pathogenicity. Vet Microbiol 208:150–158
Vreman S, McCaffrey J, Popma-de Graaf DJ, Nauwynck H, Savelkoul HFJ, Moore A, Rebel JMJ, Stockhofe-Zurwieden N (2019) Toll-like receptor agonists as adjuvants for inactivated porcine reproductive and respiratory syndrome virus (PRRSV) vaccine. Vet Immunol Immunopathol 212:27–37
Jeong J, Kang I, Kim S, Park SJ, Park KH, Oh T, Yang S, Chae C (2018) A modified-live porcine reproductive and respiratory syndrome virus (PRRSV)-1 vaccine protects late-term pregnancy gilts against heterologous PRRSV-1 but not PRRSV-2 challenge. Transbound Emerg Dis 65:1227–1234
Cruz JL, Zuniga S, Becares M, Sola I, Ceriani JE, Juanola S, Plana J, Enjuanes L (2010) Vectored vaccines to protect against PRRSV. Virus Res 154:150–160
Cui J, O’Connell CM, Costa A, Pan Y, Smyth JA, Verardi PH, Burgess DJ, Van Kruiningen HJ, Garmendia AE (2019) A PRRSV GP5-Mosaic vaccine: protection of pigs from challenge and ex vivo detection of IFNgamma responses against several genotype 2 strains. PLoS One 14:e0208801
Oh T, Kim H, Park KH, Jeong J, Kang I, Yang S, Chae C (2019) Effectiveness of a commercial porcine reproductive and respiratory syndrome virus (PRRSV) subunit vaccine against heterologous PRRSV-1 and PRRSV-2 challenge in late-term pregnant gilts. Can J Vet Res 83:248–254
Renukaradhya GJ, Meng XJ, Calvert JG, Roof M, Lager KM (2015) Inactivated and subunit vaccines against porcine reproductive and respiratory syndrome: current status and future direction. Vaccine 33:3065–3072
Murtaugh MP, Genzow M (2011) Immunological solutions for treatment and prevention of porcine reproductive and respiratory syndrome (PRRS). Vaccine 29:8192–8204
Charerntantanakul W (2012) Porcine reproductive and respiratory syndrome virus vaccines: Immunogenicity, efficacy and safety aspects. World J Virol 1:23–30
Zhang M, Wu Q, Chen Y, Duan M, Tian G, Deng X, Sun Y, Zhou T, Zhang G, Chen W, Chen J (2018) Inhibition of proanthocyanidin A2 on porcine reproductive and respiratory syndrome virus replication in vitro. PLoS ONE 13:e0193309
Li L, Tian X, Chen J, Li P, Zheng Q, Hou J (2018) Griffithsin inhibits porcine reproductive and respiratory syndrome virus infection in vitro. Arch Virol 163:3317–3325
Ge M, Xiao Y, Chen H, Luo F, Du G, Zeng F (2018) Multiple antiviral approaches of (-)-epigallocatechin-3-gallate (EGCG) against porcine reproductive and respiratory syndrome virus infection in vitro. Antiviral Res 158:52–62
Wang X, Dong W, Zhang X, Zhu Z, Chen Y, Liu X, Guo C (2021) antiviral mechanism of tea polyphenols against porcine reproductive and respiratory syndrome virus. Pathogens 10:202
Xu Z, Huang M, Xia Y, Peng P, Zhang Y, Zheng S, Wang X, Xue C, Cao Y (2021) Emodin from Aloe inhibits Porcine reproductive and respiratory syndrome virus via Toll-like receptor 3 activation. Viruses 13:1243
Chen X, Bai J, Liu X, Song Z, Zhang Q, Wang X, Jiang P (2018) Nsp1α of porcine reproductive and respiratory syndrome virus strain BB0907 impairs the function of monocyte-derived dendritic cells via the release of soluble CD83. J Virol 92:e00366-18
PubChem database. https://pubchem.ncbi.nlm.nih.gov. Accessed 3 Feb 2021
SwissTargetPrediction. http://www.swisstargetprediction.ch. Accessed 3 Feb 2021
Daina A, Michielin O, Zoete V (2019) SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 47:W357–W364
SWISS-MODEL. https://swissmodel.expasy.org. Accessed 5 Feb 2021
SAVES v6.0. https://saves.mbi.ucla.edu. Accessed 5 Feb 2021
ProSA-web. https://prosa.services.came.sbg.ac.at/prosa.php. Accessed 5 Feb 2021
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:W407-410
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Abraham MJ, Murtola T, Schulz R, Páll S, C. Smith J, Hess B, Lindahl E, (2015) GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1:19–25
Ahammad F, Alam R, Mahmud R, Akhter S, Talukder EK, Tonmoy AM, Fahim S, Al-Ghamdi K, Samad A, Qadri I (2021) Pharmacoinformatics and molecular dynamics simulation-based phytochemical screening of neem plant (Azadiractha indica) against human cancer by targeting MCM7 protein. Brief Bioinform 22:bbab098
Zheng Y, Li S, Song K, Ye J, Li W, Zhong Y, Feng Z, Liang S, Cai Z, Xu K (2022) A broad antiviral strategy: inhibitors of human DHODH pave the way for host-targeting antivirals against emerging and re-emerging viruses. Viruses 14:928
Amadori M, Listorti V, Razzuoli E (2021) Reappraisal of PRRS immune control strategies: the way forward. Pathogens 10:1073
Rusai K, Schmaderer C, Baumann M, Chmielewski S, Prokai A, Kis E, Szabo AJ, Leban J, Doblhofer R, Ammendola A, Lutz J, Heemann U (2012) Immunosuppression with 4SC-101, a novel inhibitor of dihydroorotate dehydrogenase, in a rat model of renal transplantation. Transplantation 93:1101–1107
Fitzpatrick LR, Small JS, Doblhofer R, Ammendola A (2012) Vidofludimus inhibits colonic interleukin-17 and improves hapten-induced colitis in rats by a unique dual mode of action. J Pharmacol Exp Ther 342:850–860
Hahn F, Wangen C, Hage S, Peter AS, Dobler G, Hurst B, Julander J, Fuchs J, Ruzsics Z, Uberla K, Jack HM, Ptak R, Muehler A, Groppel M, Vitt D, Peelen E, Kohlhof H, Marschall M (2020) IMU-838, a developmental DHODH inhibitor in phase II for autoimmune disease, shows anti-SARS-CoV-2 and broad-spectrum antiviral efficacy in vitro. Viruses 12:1394
Kulkarni OP, Sayyed SG, Kantner C, Ryu M, Schnurr M, Sardy M, Leban J, Jankowsky R, Ammendola A, Doblhofer R, Anders HJ (2010) 4SC-101, a novel small molecule dihydroorotate dehydrogenase inhibitor, suppresses systemic lupus erythematosus in MRL-(Fas)lpr mice. Am J Pathol 176:2840–2847
Muehler A, Peelen E, Kohlhof H, Groppel M, Vitt D (2020) Vidofludimus calcium, a next generation DHODH inhibitor for the Treatment of relapsing-remitting multiple sclerosis. Mult Scler Relat Disord 43:102129
Tang H, Liu Y, Ren R, Liu Y, He Y, Qi Z, Peng H, Zhao P (2022) Identification of clinical candidates against West Nile virus by activity screening in vitro and effect evaluation in vivo. J Med Virol 94:4918–4925
Hoffmann HH, Kunz A, Simon VA, Palese P, Shaw ML (2011) Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc Natl Acad Sci U S A 108:5777–5782
Yang CF, Gopula B, Liang JJ, Li JK, Chen SY, Lee YL, Chen CS, Lin YL (2018) Novel AR-12 derivatives, P12–23 and P12–34, inhibit flavivirus replication by blocking host de novo pyrimidine biosynthesis. Emerg Microbes Infect 7:187
Marschall M, Niemann I, Kosulin K, Bootz A, Wagner S, Dobner T, Herz T, Kramer B, Leban J, Vitt D, Stamminger T, Hutterer C, Strobl S (2013) Assessment of drug candidates for broad-spectrum antiviral therapy targeting cellular pyrimidine biosynthesis. Antiviral Res 100:640–648
Cheung NN, Lai KK, Dai J, Kok KH, Chen H, Chan KH, Yuen KY, Kao RYT (2017) Broad-spectrum inhibition of common respiratory RNA viruses by a pyrimidine synthesis inhibitor with involvement of the host antiviral response. J Gen Virol 98:946–954
Guo C, Zhu Z, Guo Y, Wang X, Yu P, Xiao S, Chen Y, Cao Y, Liu X (2017) Heparanase upregulation contributes to porcine reproductive and respiratory syndrome virus release. J Virol 91:e00625-17
Wang R, Wang X, Ni B, Huan CC, Wu JQ, Wen LB, Liao Y, Tong GZ, Ding C, Fan HJ, Mao X (2016) Syndecan-4, a PRRSV attachment factor, mediates PRRSV entry through its interaction with EGFR. Biochem Biophys Res Commun 475:230–237
Burkard C, Lillico SG, Reid E, Jackson B, Mileham AJ, Ait-Ali T, Whitelaw CB, Archibald AL (2017) Precision engineering for PRRSV resistance in pigs: macrophages from genome edited pigs lacking CD163 SRCR5 domain are fully resistant to both PRRSV genotypes while maintaining biological function. PLoS Pathog 13:e1006206
Acknowledgements
The authors wish to thank Miss. Mengyao Wang for her generous support and encouragement in experimental execution and manuscript language optimization. In addition, the critical and helpful comments from the reviewers are highly appreciated.
Funding
This work was supported by the National Natural Science Foundation (32230103), the Earmarked Fund for CARS-35, the Jiangsu Independent Innovation Fund Project (CX (22) 1006), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: YY and PJ. Methodology: YY, XL, YS, and PJ. Experiment operation: YY. Supervision: JB and PJ. Data analysis: YY, XL, YG, YS, LZ, and PJ. Writing—original draft: YY. Writing—review and editing: YY, YG, and PJ. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The experiments were conducted according to approved guidelines. All experimental protocols for animals were conducted following the National Guidelines for Housing and Care of Laboratory Animals (China) and performed in accordance with Nanjing Agricultural University (NAU) institutional regulations after approval by the Institutional Animal Care and Ethics Committee of NAU (SYXK(SU)2017-0007).
Competing interests
The authors declare that they have no competing interests.
Additional information
Handling editor: Stéphane Biacchesi.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Validation of the 3D structure of
chlorocebus sabaeus DHODH (chloDHODH). A The Ramachandran plot statistics represent the most favorable, additional allowed, generously allowed, and disallowed region with a percentage of 94.1, 5.9, 0, and 0%, respectively. B Z-score of chloDHODH is -9.57.
Additional file 2: Structure analysis of DHODHs and docking. A
Validation of the 3D structure of sus scrofa DHODH (susDHODH). B Comparative analysis of rat DHODH (ratDHODH, PDB: 1UUO), chloDHODH, and susDHODH structures by PyMOL. The structures of ratDHODH, chloDHODH, and susDHODH are labeled cyan, green, and purple, respectively. C Docked conformations of Vi with ratDHODH and susDHODH. The compound Vi is colored orange. The protein ratDHODH is colored cyan, susDHODH is colored purple, and the binding sites are shown as cavity structures. The binding energy of the Vi-ratDHODH or Vi-susDHODH complex, calculated using Autodock, is marked with an asterisk.
Additional file 3: Vidofludimus effect on HSPG2, Sdc-4, CD163 and HPSE mRNA production.
Marc-145 cells were treated with 10 µM Vi for 4, 8, 16, and 24 h, followed by qRT-PCR for HPSE (A), HSPG2 (B), Sdc-4 (C), and CD163 (D) mRNA levels. The results are from one of three independent experiments. Error bars represent the mean ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001; ns: not significant.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yang, Y., Gao, Y., Zhang, L. et al. Vidofludimus inhibits porcine reproductive and respiratory syndrome virus infection by targeting dihydroorotate dehydrogenase. Vet Res 54, 124 (2023). https://doi.org/10.1186/s13567-023-01251-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13567-023-01251-0