Abstract
Background
Radiomics-based image metrics are not used in the clinic despite the rapidly growing literature. We selected eight promising radiomic features and validated their value in decoding lung cancer heterogeneity.
Methods
CT images of 236 lung cancer patients were obtained from three different institutes, whereupon radiomic features were extracted according to a standardized procedure. The predictive value for patient long-term prognosis and association with routinely used semantic, genetic (e.g., epidermal growth factor receptor (EGFR)), and histopathological cancer profiles were validated. Feature measurement reproducibility was assessed.
Results
All eight selected features were robust across repeat scans (intraclass coefficient range: 0.81–0.99), and were associated with at least one of the cancer profiles: prognostic, semantic, genetic, and histopathological. For instance, “kurtosis” had a high predictive value of early death (AUC at first year: 0.70–0.75 in two independent cohorts), negative association with histopathological grade (Spearman’s r: − 0.30), and altered expression levels regarding EGFR mutation and semantic characteristics (solid intensity, spiculated shape, juxtapleural location, and pleura tag; all p < 0.05). Combined as a radiomic score, the features had a higher area under curve for predicting 5-year survival (train: 0.855, test: 0.780, external validation: 0.760) than routine characteristics (0.733, 0.622, 0.613, respectively), and a better capability in patient death risk stratification (hazard ratio: 5.828, 95% confidence interval: 2.915–11.561) than histopathological staging and grading.
Conclusions
We highlighted the clinical value of radiomic features. Following confirmation, these features may change the way in which we approach CT imaging and improve the individualized care of lung cancer patients.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Key points
-
Radiomics-based image features are not used in the clinic despite growing literature.
-
We selected eight features that have been informative in lung cancer screening.
-
They show additional value in revealing semantic, genetic, histopathological and prognosis heterogeneity.
-
These features are also robust to repeat imaging, segmentation operator and algorithm.
-
The findings provide new insights into individualized care of lung cancer patients.
Background
More than one in ten human cancers occurs in the lung [1]. The biological, spatial, and temporal heterogeneity of lung cancer makes its clinical management a critical challenge [2]. Computed tomography (CT) provides a means to observe the lung noninvasively and visualize lesions macroscopically, which can be performed repeatedly when necessary; thus, CT is core to the diagnosis and treatment workflow of lung cancer. The advantage of the knowledge inherent in CT images, however, is not being fully applied in the clinic, as only a few semantic characteristics or simplistic metrics (e.g., diameter) are routinely used [3, 4]. In the research field, efforts have been made in the translation of images into quantitative features that describe tumor shape and texture characteristics, as well as the association of these features with known clinical endpoints. This approach, termed radiomics, provides a unique opportunity for the generation of innovative methods for phenotypic profiling of lung cancer [5]. Several studies have demonstrated the potential use of radiomics-based metrics to deepen our knowledge on how lung cancers differ from benign lesions [6], and how cancer types differ from one another regarding development [7], prognosis [8], treatment response, and recurrence [9].
There are several important barriers that impede the use of radiomic features clinically: (1) the majority of radiomic research reports on isolated results regarding specific tasks, such as cancer diagnosis or prognosis; thus far, the value of radiomic features has rarely been demonstrated in independent datasets and for multiple tasks [10, 11]; (2) many radiomic features are computationally complex and difficult to interpret; lack of consensus on the definition [12] and interpretation of the associations between specific radiomic features with known cancer phenotypes make it challenging for physicians to uptake [3]; and (3) considering the variations in imaging acquisition and processing, the percentage of radiomic features that are deemed reproducible is low, about 6–43% in phantom studies and 10% in patient studies [13].
Because it has been ~ 10 years since radiomics gained its name, it is a suitable time to consider the controversies between a rapidly growing body of publications and the clinical application [14]. In a previous proof-of-concept study, in which we determined the optimal time for diagnostic testing in lung cancer screening, we found that several radiomic features were robust to image noise, associated with semantic characteristics, and predictive of lung cancer diagnosis [15]. In the present study, we validate the clinical value of these radiomic features based on their ability to predict long-term prognosis of lung cancer patients. We also provide additional support for their clinical uptake by showing the associations of these features with semantic, genetic, and histopathological heterogeneity in lung cancer, as well as the measurement reliability of these features.
Methods
Data sources
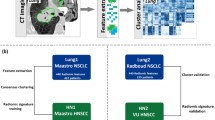
The proof-of-concept study was conducted between 2014 and 2019 at Peking Union Medical College Hospital and included 92 patients enrolled in a lung cancer screening program [15]. The CT images used in this study were from 236 lung cancer patients at three other institutes [16, 17]. Cohort A: a total of 146 early-stage non-small cell lung cancer (NSCLC) patients were recruited between years 2008 and 2012 at Stanford University School of Medicine and underwent CT and/or positron emission tomography (PET)/CT scan before surgical treatment (three patients had no CT images and were excluded). Cohort B: CT images of 61 lung adenocarcinoma patients before surgical treatment were acquired between years 2006 and 2009 at H. Lee Moffitt Cancer Center. Cohort C: CT images were collected in 2017 at Memorial Sloan-Kettering Cancer Center and were from 32 patients with NSCLC who underwent two CT scans of the chest within 15 min.
The images were retrieved from the Cancer Imaging Archive (https://www.cancerimagingarchive.net). To reflect the diverse clinical settings of each clinical center, there was no attempt to harmonize the image acquisition protocols (the clinical settings of each center are summarized in Appendix Table 3). The data usage policy for the collection of each image was followed. There was no need for ethical approval because all the patient information was de-identified.
Image segmentation and feature extraction
The process of image segmentation and feature extraction has been previously standardized [15] and was performed accordingly in this study.
In brief, regions-of-interest (ROIs) were delineated following an interactive process: cross-sectional slices with the maximum area per pulmonary lesion were selected, and the threshold method (for juxtapleural lesions or part-solid and non-solid lesions) or manual initial contouring (for complex cases) was exercised, followed by automated refinement using Image Segmenter Toolbox, MATLAB R2018a until visual satisfaction. Two investigators contoured the ROIs on the initial scan images to assess the between-operator variation, and one of the investigators used two segmentation algorithms—region-based and edge-based active contour models—to assess the between-algorithm variation.
Radiomic features were then extracted from the ROIs using online and in-house adapted codes of MATLAB software (see [15] for specific definitions). We evaluated two sets of radiomic features: (1) eight selected features, including a shape feature (circularity; quantifying the degree of the ROI approximating a circle), three statistical features (variance, kurtosis, and energy; quantifying the dispersion, sharpness and magnitude of ROI brightness, respectively), three texture features (cluster-shade, maximum-probability, and long-run high gray-level emphasis mean [LongHEM]; quantifying the asymmetry, predominance of coexisting image pixel pairs, or the degree of bright coarse structural textures, respectively), and a wavelet feature (long-run emphasis mean on approximation signal; reflecting the structural texture in a probably finer resolution); these features had undergone a rigorous selection process in the proof-of-concept study using the following criteria: robustness to artificial image noise, predictive performance of cancer diagnosis, and non-redundancy (i.e., small effect of collinearity between features); see [15] for detailed feature selection workflow; (2) 11 features that revealed similar clinical utility potential but correlated to one of the selected features. We present the eight selected features in the results section, and the remaining information is available in Appendix.
For comparison purposes, diameter was calculated as a classical image metric, which was calculated by the average of the major and minor axis lengths, rounded to the nearest integer; semantic characteristics of the cancer images (including solid, lobular, specular, juxtapleural, and pleura tags) were interpreted by a researcher and a radiologist experienced at reading chest CT images.
Analytic workflow and methods
Data on survival outcomes were available from Cohorts A and B, and we used these two cohorts for examining the prognostic value of the radiomic features. We randomly divided Cohort A (considering its relatively large sample size) using a 2:1 ratio for the training and testing groups; independent external validation was then performed using Cohort B. A machine-learning random survival forest (RSF) model was used to build a composite radiomic score that could more comprehensively evaluate the cancer characteristics than a single feature. The RSF is an extension of random forest (ensemble of tree models) to survival outcome [18] and was implemented with R package “randomForestSRC,” with hyper-parameters (ntree = 50, nodesize = 10, nodedepth = 4) determined by grid search. The predicted risk for each patient was rescaled to 0–100 by normalization to convert it to a score. Prognosis values of the radiomic features and score were measured by C-statistic (overall discrimination of time-to-survival outcome), as well as at several points of interest (e.g., to determine 2- or 5-year survival), using time-dependent area under the receiver operating curve (AUCt). Hazard ratio (HR) of death was computed between groups of patients with high and low scores.
To determine a clinical explanation of the novel features, in the proof-of-concept cohort, we associated the radiomic features with the semantic characteristics that are widely used in lung cancer image interpretation [15]; we further validated these associations in Cohorts A, B, and C. We also used information available in Cohort A on gene mutations (epidermal growth factor receptor [EGFR], Kirsten rat sarcoma viral oncogene homolog [KRAS], and anaplastic lymphoma kinase [ALK]) and histopathology (types and grades) to confirm the findings biologically. The associations of the radiomic features with these micro-level subtypes were examined using a differential expression analysis approach using the Wilcoxon test and Spearman’s correlation.
Lastly, for comprehensive assessment of the measurement reliability of the radiomic features, we examined feature reproducibility in terms of image acquisition (between-repeat scans) and segmentation (between operators and between algorithms) using data from Cohort C. Features with an intraclass coefficient (ICC) ≥ 0.8 were considered robust to the abovementioned variation. Bland–Altman plots were drawn for visual analysis.
The statistical tests were two-sided, with a significance level of 0.05. All the statistical analyses were performed with R version 3.5.2.
Results
Patient characteristics
Across the three patient cohorts (Table 1), the characteristics of lung cancer did not vary significantly regarding location (31.3–40.6% in the left lung [p = 0.2765]) or semantic characteristics (65.6–68.9% were a lobular shape, 24.5–37.5% were a spiculated shape, 37.7–59.4% had pleural invasion [elastic, visceral, or parietal], and 15.6–23.0% had pleura tags [all p > 0.050]), with the exception of image intensity, which varied between cohorts (50.8–81.3%, solid intensity [p = 0.0155]). Between patients in Cohorts A and B, there were no significant differences regarding cancer stage: 17.5% vs 29.3% staged IIIA or above, respectively; or regarding survival: median survival time was 40.0 vs 62.8 months, respectively (both p > 0.050).
Prognosis value
When used alone, the feature, “LongHEM,” showed a prognostic value that was equal to diameter. The overall C-statistic was 0.627 (LongHEM) vs 0.570 (diameter) in Cohort A, and 0.602 vs 0.605 in Cohort B; AUCt for predicting survival beyond 2 and 5 years was 0.617 (LongHEM) vs 0.612 (diameter) and 0.588 vs 0.597, respectively, in Cohort A, and 0.648 vs 0.618 and 0.670 vs 0.678, respectively, in Cohort B. A similar prognostic value was observed regarding features “kurtosis,” “energy,” and “maximum-probability” (Appendix Table 4).
Next, we developed a composite score using the eight selected features. The prognostic value of the composite score (Appendix Table 5) was greater than diameter, the five semantic characteristics, and a combination of these routine characteristics. For instance, when used for predicting survival beyond 5 years, the AUCt was 0.855 (train), 0.780 (test), and 0.760 (external validation) for the composite score, vs 0.733 (train), 0.622 (test), and 0.613 (external validation) for a combination of diameter and semantic characteristics.
When patients in Cohorts A and B were stratified according to their median scores, we found significantly different prognosis between groups with low and high scores (p < 0.050 in both cohorts; Fig. 1A, B). Patients with a low radiomic score were associated with higher chances of survival from the beginning (1 year) of the follow-up process, and the between-group divergence in survival curves became more apparent after this time point. The performance of the radiomic score was better compared to histopathological staging (Fig. 1C, D) and grading (Fig. 1E; data only available for Cohort A). The HR of patients with a high vs low radiomic score was 5.828 (95% confidence interval [CI] 2.915–11.561) in Cohort A and 2.722 (95% CI 1.117–6.633) in Cohort B. The prognostic value was demonstrated across age, gender, and smoking category subgroups (Fig. 1F), and more pronounced among older (aged ≥ 70 years, HR 8.189), female (HR 7.210), and non-smoking patients (HR 15.190; all p < 0.050 against a null effect).
Survival of lung cancer patients. Stratified by composite radiomic score, (A) Cohort A, (B) Cohort B; histopathological staging, (C) Cohort A, (D) Cohort B; histopathological grading, (E) Cohort A (data not available for Cohort B); and by demographic subgroups for the examination of score value, (F) Cohort A
Associations with semantic, genetic, and histopathological profiles
First, by associating the radiomic features with the semantic characteristics (Table 2), the following findings in the proof-of-concept cohort were independently confirmed in Cohort A: up-regulation of feature “maximum-probability” in cancers of solid intensity, of feature “variance” in cancers of spiculated shape, and of features “circularity” and “energy” in cancers attached to the pleura. We also found an association between “circularity” and juxtapleural location in Cohort B. Additionally, down-regulation of the feature “kurtosis” in juxtapleural cancers was demonstrated in Cohorts A and B, and up-regulation of feature “LongHEM” in juxtapleural cancers was observed in Cohorts A and C. The directions of the significant differentially expressed features were consistent across different cohorts, with the exception of “LongHEM” and spiculated shape.
We then analyzed the relatively large sample size and the available information from Cohort A to evaluate radiomic expression patterns regarding different genetic and histopathological profiles.
EGFR-mutated (n = 23) relative to wild-type (n = 93) lung cancers showed an up-regulation of the feature “kurtosis” (median: 3.87 vs 2.48; p = 0.0238), and down-regulation of the features “maximum-probability” (0.53 vs 0.75; p = 0.0130) and “energy” (0.05 vs 0.09; p = 0.0401; Fig. 2A). Moreover, a down-regulation of feature “variance” was observed in ALK-translocated (n = 2) vs wild-type (n = 109) cancers (4.60 vs 8.64; p = 0.0282), but there was no differentially expressed features regarding KRAS-mutated (n = 27) vs wild-type (n = 88) cancers.
Association of image metrics with gene and histopathological phenotypes. A Volcano plot showing the significantly up-regulated (red) and down-regulated (blue) image metrics (p < 0.050) regarding EGRF mutation. B Radar plot of differentially expressed image metrics regarding histopathological type. C Dose–response plot of histopathological grade with image metrics. *p < 0.050; **p < 0.010. EGFR: epidermal growth factor receptor
For cancers stratified by different histopathological groups (adenocarcinoma, squamous cell lung cancers, and other NSCLCs; Fig. 2B), the difference observed for the feature “variance” was highly significant (8.25 vs 8.94 vs 9.61; p = 0.0031). Other differentially expressed features regarding these histopathological groups included “cluster-shade” and “maximum-probability” (p < 0.050 for both of the features). Moreover, we found that four features significantly increased or decreased along with an increase in histopathological grade (Fig. 2C): “LongHEM” (Spearman’s coefficient of correlation: 0.331), “kurtosis” (− 0.329), “energy” (0.325), and “maximum-probability” (0.281; all p < 0.010).
Compared to the selected radiomic features, diameter had no discriminative power regarding cancers with EGFR or ALK mutations, nor for histopathological types (all p > 0.050). The correlation with histopathological grade (Spearman’s coefficient of correlation: 0.226; p < 0.010) was weaker than for the four aforementioned radiomic features (“LongHEM,” “kurtosis,” “energy,” and “maximum-probability”).
Feature reproducibility
Despite the visual differences in the segmented ROIs between repeat scans and segmentation operators and algorithms (see Fig. 3 as an example), the eight radiomic features were generally not affected by these sources of variation (Fig. 4; Appendix Table 6). Specifically, ICC ranged from 0.81 (“LongHEM”) to 0.98 (“energy”) in repeat scans, and from 0.80 (“circularity”) to 0.99 (“kurtosis”) between segmentation operators and algorithms, with the exception of an ICC of 0.70, which was observed for between operators, and an ICC of 0.57, which was observed for between algorithms (“cluster-shade”).
Discussion
Only a few quantitative or semantic features are routinely used in lung cancer image interpretation, and these features are considered insufficient because they are too simple or prone to inter-observer variability [19]. Currently, despite rapidly expanding literature on radiomics, there is no clinical use of radiomic features [20]. On the basis of our previous report [15] and the findings in the current study, we show that the eight selected radiomic features are predictive of both the diagnosis and prognosis of lung cancer, descriptive of semantic characteristics, and some are indicative of genetic and histopathological profiles. The selected features were largely robust to variation in image noise, repeat imaging, and visual differences in ROIs caused by segmentation operators and algorithms. When combined, the radiomic features showed a moderate prognosis value (AUCt for 5-year survival: 0.760 in external validation) and capability for risk stratification (HR 5.828 and 2.722 in two independent cohorts; the HRs among elderly, female, and non-smoking subgroups were 8.189, 7.210, and 15.190, respectively), and the stratification capability was even better than histopathological staging and grading. These results highlight the feasibility of using new metrics for lung cancer images in the clinical setting.
Because of the substantial heterogeneity in lung cancers, new methods for subtyping are needed. This issue has led to numerous efforts regarding cancer characterization at the tissue (imaging), cellular (microcopy), and molecular (genetic test) levels [5, 21]. Of these methods, histopathological methods are some of the oldest methods (specialty of histopathology techniques dates back to 1838 [22]), which laid a foundation for the diagnosis and treatment of cancer. Alterations to the cancer classification system are being pursued in the age of precision medicine [21], driven by improved methods that are able to reveal the molecular basis of the disease. The time for the introduction of medical imaging lied in between [9]; strengthened by a better computation power, description of the cancer morphology (and potentially its evolving biology) can go beyond human perceptions [23]. This may help to explain why the radiomic score outperformed the histopathological staging and grading in this study. Similar findings were observed in another study [24] that used more features for the construction of a radiomic signature than our study. Our finding on the interweaving associations between radiomic features and pathohistological and genetic profiles, as well as findings from several other publications (regarding prediction of lung cancer histopathology [25], EGFR and ALK mutations [26, 27], and prognosis among ALK-positive patients [28] using radiomic features), altogether confirm the presence of a connection between the macro-level imaging information and micro-level biology. As CT images are routinely collected, the analysis of radiomic features could be a relatively inexpensive and non-invasive means for cancer profiling.
Radiomic features are prone to variations in each step, from image acquisition to segmentation [29]. This is a key issue for the application of radiomics. For instance, many of the radiomic-semantic associations observed in our proof-of-concept study, validated in Cohort A or B, were not found in Cohort C. Despite the small sample size in Cohort C, we believe this discrepancy was due to the larger slice thickness (2.5–6.0 mm) in the samples in Cohort C compared with the other cohorts (0.6–3.0 mm). In a recent publication [13], slice thickness had the largest impact on feature reproducibility compared with other factors; slice thickness impacted visualization and interpretation of semantic characteristics (e.g., spiculated shape) according to our experience. We did not focus the present study on volumetric radiomic features or features requiring sophisticated quantification (such as those based on wavelet and other transformed images) because multiple previous publications [30, 31] and our proof-of-concept study [15] showed less reproducibility for these features than for shape, statistical, and a subset of texture features, even for peritumoral radiomics [31]. Instead, in the present study, following a rigorous selection process, we focused on several radiomic features. The feature reproducibility results confirmed that these features were reliable. Therefore, the analysis of these features may help to avoid spurious findings in subsequent analyses and in the clinical setting.
Among the selected radiomic features, “kurtosis” is a simple statistical feature describing the sharpness of the image intensity level distribution. Because of its simplicity, features of this category may be more robust than measuring shape and texture features (e.g., “circularity” and “cluster-shade” in this study), according to a systematic review [29]. For instance, “kurtosis” showed an ICC of > 0.96 between repeat scans and between segmentation operators and algorithms, which is relatively higher compared with other selected features. The clinical potential of the “kurtosis” feature as a new image metric for classification and progression of lung diseases has been repeatedly observed [32]. We further revealed several good properties of “kurtosis,” such as its high predictive value of early death (AUC at first year: 0.70–0.75 in two independent cohorts), negative linear association with histopathological grade (Spearman’s r: − 0.30), and significantly altered levels regarding EGFR mutations and regarding nearly all the semantic characteristics investigated. On the basis of these findings, we expect a wider application of this quantitative metric in the future given its ease-of-computation and interpretation.
Nevertheless, we could not expect too much from a single metric as a solution to a specific clinical task. For instance, in this study, the prognostic value of the radiomic features were, at best, equal to the use of diameter when used alone, and for most of the features, such values may vary over time. For some features, the temporal trend may also differ between cohorts (Appendix Table 4). Owing to their substantial heterogeneity, lung cancers of the same histopathological type can show varying imaging and survival characteristics; such variations have also been reported in cancers of the same genetic type [33]. Implications of these complex findings are far-reaching. First, these findings indicate that a lot more progress required in the pursuit for novel methods for cancer characterization. Regarding morphology, it was recently indicated that homology-based [34], peritumoral [35], or sequential radiomic features [36] may add to the value of standard radiomics, though further validations are needed. Second, to capture and differentiate tumoral heterogeneity, we need to take advantage of the effect of combined approaches. One example of this from the current study is that when the selected features (non-redundant measurements of cancer appearance) were combined as a score, their prognostic value was significantly enhanced. Therefore, mathematical formulas, statistical models, or machine learning algorithms deserve further investigation in this context [37]. Third, despite our enthusiasm for optimizing models’ precision for precise cancer care, we argue for an equal emphasis on basic techniques that are highly reproducible and allow adequate interpretation of the results.
There are several limitations in this study: (1) although digital images can be stored over long periods, the retrospective nature of this study limited our access to the demographic and genetic data of some patients; (2) CT images from more diverse scanning protocols should be considered for a better extrapolation of the results, and the impact of varying scanning protocols on feature reproducibility remain inadequately assessed; (3) the use of PET/CT images can improve the accuracy of cancer staging; therefore, it remains unknown whether the results on the prognosis value of the composite radiomic score against histopathological staging may be altered by adding such information; (4) we only examined a small number of selected radiomic features and applied them to a case-mix of lung cancer patients; this may have led to a lower prognostic accuracy compared with other reports [38, 39]; (5) the value of the selected radiomic features was only assessed at a certain time point (before surgery). It was indicated in our previous study that features such as “LongHEM” may be a good metric for monitoring purposes when measured repeatedly (i.e., delta radiomics); validation of this finding requires longitudinal data and further analysis.
Conclusion
Following a previous work, we demonstrated in this study that several selected radiomic features are predictive of survival outcome and can differentiate between semantic, genetic, and histopathological heterogeneities in lung cancer. Moreover, the measurement of these features was reproducible in repeat scans and image segmentation. Following confirmation, the novel features described in this study may help to improve approaches of CT image analysis, and therefore, may improve individualized care of lung cancer patients.
Availability of data and materials
The datasets supporting the conclusions of this article are available in the Cancer Imaging Archive repository: https://doi.org/10.7937/K9/TCIA.2017.7hs46erv. https://wiki.cancerimagingarchive.net/display/Public/NSCLC+Radiogenomics; https://doi.org/10.7937/K9/TCIA.2015.A6V7JIWX. https://wiki.cancerimagingarchive.net/display/Public/LungCT-Diagnosis; https://doi.org/10.7937/K9/TCIA.2015.U1X8A5NR. https://wiki.cancerimagingarchive.net/display/Public/RIDER+Lung+CT.
Abbreviations
- ALK:
-
Anaplastic lymphoma kinase
- AUCt:
-
Time-dependent area under the receiver operating curve
- CI:
-
Confidence interval
- CT:
-
Computed tomography
- EGFR:
-
Epidermal growth factor receptor
- HR:
-
Hazard ratio
- ICC:
-
Intraclass coefficient
- IQR:
-
Inter-quartile range
- KRAS:
-
Kirsten rat sarcoma viral oncogene homolog
- LongHEM:
-
Long-run high gray-level emphasis mean
- NSCLC:
-
Non-small cell lung cancer
- PET:
-
Positron emission tomography
- ROI:
-
Region-of-interest
References
International Agency for Research on Cancer. Cancer today. Available: https://gco.iarc.fr/today/home. Accessed 15 Sept 2021
Zhao C, Fillmore CM, Hammerman PS et al (2014) Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer 14:535–546
Aerts HJ (2016) The potential of radiomic-based phenotyping in precision medicine: a review. JAMA Oncol 2:1636–1642
Constanzo J, Wei L, Tseng HH et al (2017) Radiomics in precision medicine for lung cancer. Transl Lung Cancer Res 6:635–647
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Khawaja A, Bartholmai BJ, Rajagopalan S et al (2020) Do we need to see to believe? Radiomics for lung nodule classification and lung cancer risk stratification. J Thorac Dis 12:3303–3316
Tan M, Ma W, Sun Y et al (2021) Prediction of the growth rate of early-stage lung adenocarcinoma by radiomics. Front Oncol 11:658138
Aerts HJ, Velazquez ER, Leijenaar RT et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Rogers W, Thulasi Seetha S, Refaee TAG et al (2020) Radiomics: from qualitative to quantitative imaging. Br J Radiol 93:20190948
Timmeren V, Janna E, Elmpt V et al (2019) Longitudinal radiomics of cone-beam CT images from non-small cell lung cancer patients: evaluation of the added prognostic value for overall survival and locoregional recurrence. Radiother Oncol 136:78–85
Kim DW, Jang HY, Kim KW et al (2019) Design characteristics of studies reporting the performance of artificial intelligence algorithms for diagnostic analysis of medical images: results from recently published papers. Korean J Radiol 20:405–410
Zwanenburg A, Vallières M, Abdalah MA et al (2020) The image biomarker standardization initiative: standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 295:191145
Meyer M, Ronald J, Vernuccio F et al (2019) Reproducibility of CT radiomic features within the same patient: influence of radiation dose and CT reconstruction settings. Radiology 293:190928
Doran SJ, Kumar S, Orton M et al (2021) “Real-world” radiomics from multi-vendor MRI: an original retrospective study on the prediction of nodal status and disease survival in breast cancer, as an exemplar to promote discussion of the wider issues. Cancer Imaging 21:37
Wang Z, Li N, Zheng F et al (2021) Optimizing the timing of diagnostic testing after positive findings in lung cancer screening: a proof of concept radiomics study. J Transl Med 4(19):191
Bakr S, Gevaert O, Echegaray S et al (2017) NSCLC radiogenomics collection. Cancer Imaging Arch. https://doi.org/10.7937/K9/TCIA.2017.7hs46erv
Grove O, Berglund AE, Schabath MB et al (2015) Quantitative computed tomographic descriptors associate tumor shape complexity and intratumor heterogeneity with prognosis in lung adenocarcinoma. Cancer Imaging Arch. https://doi.org/10.7937/K9/TCIA.2015.A6V7JIWX
Zhao B, Schwartz, Lawrence H et al (2015) RIDER_Lung CT. The cancer imaging archive. https://doi.org/10.7937/K9/TCIA.2015.U1X8A5NR
Ishwaran H, Kogalur UB, Chen X, Minn AJ (2011) Random survival forests for high-dimensional data. Stat Anal Data Min 4:115–132
van Riel S, Jacobs C, Scholten ET et al (2019) Observer variability for lung-RADS categorisation of lung cancer screening CTs: impact on patient management. Eur Radiol 29:924–931
Zhao B (2021) Understanding sources of variation to improve the reproducibility of radiomics. Front Oncol 11:633176
de Sousa VML, Carvalho L (2018) Heterogeneity in lung cancer. Pathobiology 85:96–107
Titford M (2006) A short history of histopathology technique. J Histotechnol 29:99–110
Gillies RJ, Kinahan PE, Hricak H (2016) Radiomics: images are more than pictures, they are data. Radiology 278:563–577
D’Antonoli TA, Farchione A, Lenkowicz J et al (2020) CT radiomics signature of tumor and peritumoral lung parenchyma to predict nonsmall cell lung cancer postsurgical recurrence risk. Acad Radiol 27:497–507
Wu W, Parmar C, Grossmann P et al (2016) Exploratory study to identify radiomics classifiers for lung cancer histology. Front Oncol 6:71
Li S, Ding C, Zhang H et al (2019) Radiomics for the prediction of EGFR mutation subtypes in non-small cell lung cancer. Med Phys 46:4545–4552
Song L, Zhu Z, Mao L et al (2020) Clinical, conventional CT and radiomic feature-based machine learning models for predicting ALK rearrangement status in lung adenocarcinoma patients. Front Oncol 10:369
Li H, Zhang R, Wang S et al (2020) CT-based radiomic signature as a prognostic factor in stage IV ALK-positive non-small-cell lung cancer treated with TKI crizotinib: a proof-of-concept study. Front Oncol 10:57
Alberto T, Leonard W, Andre D et al (2018) Repeatability and reproducibility of radiomic features: a systematic review. Int J Radiat Oncol Biol Phys 102:1143–1158
Zhao B, Tan Y, Tsai WY et al (2016) Reproducibility of radiomics for deciphering tumor phenotype with imaging. Sci Rep 6:23428
Tunali I, Hall LO, Napel S et al (2019) Stability and reproducibility of computed tomography radiomic features extracted from peritumoral regions of lung cancer lesions. Med Phys 6:5075–5085
Frix AN, Cousin F, Refaee T et al (2021) Radiomics in lung diseases imaging: state-of-the-art for clinicians. J Pers Med 11:602
Lee G, Bak SH, Lee HY (2018) CT radiomics in thoracic oncology: technique and clinical applications. Nucl Med Mol Imaging 52:91–98
Kadoya N, Tanaka S, Kajikawa T et al (2020) Homology-based radiomic features for prediction of the prognosis of lung cancer based on CT-based radiomics. Med Phys 47:2197–2205
Pérez-Morales J, Tunali I, Stringfield O et al (2020) Peritumoral and intratumoral radiomic features predict survival outcomes among patients diagnosed in lung cancer screening. Sci Rep 10:10528
Liu Y, Wu M, Zhang Y et al (2021) Imaging biomarkers to predict and evaluate the effectiveness of immunotherapy in advanced non-small-cell lung cancer. Front Oncol 11:657615
Boek P, Binczyk F, Prazuch W et al (2021) Radiomics and artificial intelligence in lung cancer screening. Transl Lung Cancer Res 10:1186–1199
He B, Zhao W, Pi JY et al (2018) A biomarker basing on radiomics for the prediction of overall survival in non-small cell lung cancer patients. Respir Res 19:199
Acknowledgements
We thank all the participants, researchers and staff involved in the collection, processing and maintenance of the data, as well as the related sponsors, for their important contributions to this study.
Funding
This study was supported by the CAMS Innovation Fund for Medical Sciences (grand number: 2021–1-I2M-022) and the Fundamental Research Funds for the Central Universities (grand number: 3332021038). The funding bodies were not involved in the design, data collection, analysis, or interpretation of data, nor involved in writing the manuscript.
Author information
Authors and Affiliations
Contributions
ZW, JJ and WS conceived and designed the study; ZW, CY and WH did the analysis; XS, FZ, XX interpreted the image data; WH, FX, PW and YC interpreted the analysis and plotted the figures; ZW drafted the work and all the authors substantively revised it. All authors contributed to the writing of and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, Z., Yang, C., Han, W. et al. Quantifying lung cancer heterogeneity using novel CT features: a cross-institute study. Insights Imaging 13, 82 (2022). https://doi.org/10.1186/s13244-022-01204-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13244-022-01204-9