Abstract
Objectives
The study evaluated sub-microscopic malaria infections in pregnancy using two malaria Rapid Diagnostic Tests (mRDTs), microscopy and RT-PCR and characterized Plasmodium falciparum dihydrofolate reductase (Pfdhfr) and Plasmodium falciparum dihydropteroate synthase (Pfdhps) drug resistant markers in positive samples.
Methods
This was a cross sectional survey of 121 pregnant women. Participants were finger pricked, blood drops were collected for rapid diagnosis with P. falciparum histidine-rich protein 11 rapid diagnostic test kit and the ultra-sensitive Alere Pf malaria RDT, Blood smears for microscopy and dried blood spots on Whatman filter paper for molecular analysis were made. Real time PCR targeting the var acidic terminal sequence (varATS) gene of P. falciparum was carried out on a CFX 96 real time system thermocycler (BioRad) in discriminating malaria infections. For each run, laboratory strain of P. falciparum 3D7 and nuclease free water were used as positive and negative controls respectively. Additionally, High resolution melt analyses was employed for genotyping of the different drug resistance markers.
Results
Out of one hundred and twenty-one pregnant women sampled, the SD Bioline™ Malaria Ag P.f HRP2-based malaria rapid diagnostic test (mRDT) detected eight (0.06%) cases, the ultra-sensitive Alere™ malaria Ag P.f rapid diagnostic test mRDT had similar outcome in the same samples as detected by the HRP2-based mRDT. Microscopy and RT-PCR confirmed four out of the eight infections detected by both rapid diagnostic tests as true positive and RT-PCR further detected three false negative samples by the two mRDTs providing a sub-microscopic malaria prevalence of 3.3%. Single nucleotide polymorphism in Pfdhps gene associated with sulphadoxine resistance revealed the presence of S613 mutant genotypes in three of the seven positive isolates and isolates with mixed wild/mutant genotype at codon A613S. Furthermore, four mixed genotypes at the A581G codon were also recorded while the other Pfdhps codons (A436G, A437G and K540E) showed the presence of wild type alleles. In the Pfdhfr gene, there were mutations in 28.6%, 28.6%, and 85.7% at the I51, R59 and N108 codons respectively. Mixed wild and mutant type genotypes were also observed in 28.6% each of the N51I, and C59R codons. For the Pfcrt, two haplotypes CVMNK and CVIET were observed. The SVMNT was altogether absent. Triple mutant CVIET 1(14.3%) and triple mutant + wild genotype CVIET + CVMNK 1(14.3%) were observed. The Pfmdr1 haplotypes were single mutants YYND 1(14.3%); NFND 1(14.3%) and double mutants YFND 4(57.1%); YYDD 1(14.3%).
Similar content being viewed by others
Introduction
Nigeria contributes about 27% to the global malaria burden and 24% to malaria death globally [1]. Of the 33.2 million pregnant women in 2019, 35% (11.8 million) were exposed to malaria infection in 33 moderate to high transmission countries located in the World Health Organisation (WHO) Africa Region [1]. Pregnant women are among the high risk groups vulnerable to malaria infection due to their temporarily compromised immune system [2]. Pregnancy associated malaria (PAM) is a frequent occurrence in women living in malaria endemic countries especially in tropical and sub-tropical regions of the world such as Africa [3, 4]. PAM is defined as the detection of asexual parasite stages in peripheral or sequestered blood cells in the placenta of a pregnant woman [3]. Plasmodium falciparum is the most prevalent and implicated species in PAM and causes complications both in the mother and the foetus [5, 6]., sometimes leading to maternal anaemia, spontaneous abortion, stillbirths, premature and low birth weight [3, 6,7,8,9,10]. As a result of the presence of sub-microscopic infections in pregnant women, detection of malaria infections requires high sensitive diagnostic tools that can detect low parasite density [11]. Traditionally, microscopy is the gold standard, but due to the various challenges (inadequate training/experienced of microscopy readers, deficiency in personnels, sub-standard or inadequate equipment, lack of power supply etc.) that surrounds it, [12, 13] rapid diagnostic tool such as P. falciparum histidine rich protein II (HRP-2) malaria RDTs are deployed in many endemic areas where microscopy is unavailable. It requires less training, cost effective and produces timely result [14,15,16]. However, the circulation of parasite with deleted PfHRP2 gene makes parasite detection with mRDT difficult [17, 18] Therefore, more sensitive mRDT would be effective in providing timely results [19–20] WHO treatment policy for uncomplicated malaria in the general population is the use of artemisinin based combination therapy while in pregnant women, at least 2 doses of Intermittent Preventive Treatment with sulfadoxine-pyrimethamine (IPTp-SP) after quickening. In sub-Saharan Africa, IPTp-SP has been shown to reduce adverse infant and maternal outcomes such as maternal anaemia, low birth weight, placental malaria and perinatal mortality [21, 22], and neonatal mortality [23]. The implementation and effectiveness of this approach has been largely riddled by the development of P. falciparum resistance to SP [24, 25]. Single nucleotide polymorphisms in the P. falciparum dihydrofolate reductase (Pfdhfr) and the P. falciparum dihydropteroate synthase (Pfdhps) genes have been associated with resistance to pyrimethamine and sulfadoxine respectively. Substitutions at the Pfdhfr 51, 59, 108, 164, and Pfdhps 437, 540, 581 and 613 codons have been evaluated and detected in various populations in different endemic settings [26, 27]. Mutations at the P. falciparum multi-drug resistant gene 1 Pfmdr1) has been associated with various artemisinin partner drugs. For instance, substitutions in codons N86, 184 F and D1246 of Pfmdr1 have been associated with resistance to lumefantrine and amodiaquine while the Pfmdr1 86Y allele has been strongly linked with chloroquine and amodiaquine resistance [44, 46]. Moreover, 1246Y alleles have also been shown to increase P. falciparum susceptibility to mefloquine, halofantrine and artemisinin [45]. While the resistance of P. falciparum kelch 13 propeller domain was uncommon, there were fixation in the prevalence of Pfcrt at codons 74–76 and 86Y and high prevalence of Pfdhfr and Pfdhps among Nigeria populations [47]. The NFD haplotype (86 N-184 F– 1246D) have been reported to significantly associate with treatment failure among children under five [26] in Nigeria. However, there is paucity of data of these resistant associated mutations in pregnant women from Lagos State, Nigeria. Therefore, this study was designed to i) evaluate sub-microscopic malaria infections in pregnant women using two mRDTs, microscopy and RT-PCR; ii). Characterize Pfdfr, Pfdhps, Pfcrt and Pfmdr1 drug resistant markers in positive isolates.
Materials and methods
Study design, sites and participants
This was a cross-sectional study that involved pregnant women attending antenatal clinic in Epe and Ogudu Primary Health Centres, in Lagos State. The study sites are located in the south– western part of Nigeria and have high malaria transmission. The entomological inoculation rate as reported from a previous study [28] is 8.4 infective bites per person per month (ib/p/m) by human bait and 5.45 ib/p/m by pyrethrum sprays catch. The time frame for the collection of the pregnant women samples was from November, 2019 to March, 2021.
Written and verbal consents were obtained from all the participating pregnant women before the commencement of the study. Only consenting pregnant women with no pregnancy-related complications were enrolled into the study. Pregnant women not meeting study criteria were excluded.
After sensitisation and consenting, one hundred and twenty- one pregnant women of were recruited into the study.
Blood collection
Participants were finger -pricked and blood drops collected for rapid diagnosis with P. falciparum histidine-rich protein II rapid diagnostic test kit and the ultra-sensitive Alere Pf malaria RDT, blood smears for microscopy and dried blood spots (DBS) on Whatman filter paper for molecular analysis were also collected. DBS were air-dried and kept in sealed plastic bags containing desiccants until use.
For pregnant women that tested positive, the attention of their attending obstetrician and gynecologist were brought to the situation and care thereafter were outside the purview of the study.
DNA extraction and real time PCR (RT-PCR) for malaria molecular diagnosis
Genomic DNA of all samples was extracted from three punches of 3 mm dried blood spot using the QIAamp DNA Blood Mini Kit (Qiagen®, Hilden, Germany), eluted in a 100 µL final volume and stored at − 20 °C until use.
To accurately detect the presence of malaria parasites from these gDNA samples, RT-PCR targeting the var acidic terminal sequence (varATS) gene of P. falciparum was carried out as detailed elsewhere [29, 30]. Briefly, 5 µL of template gDNA was added to a master mix containing 1 µL of nuclease-free water, 10 µL of 2x Taqman Universal PCR Mastermix (Applied Biosystems, New Jersey, USA), 1.6 µL of 10 µM forward and reverse primer each and 0.8 µL of 10 µM probe. The master mix together with the template gDNA was run on a CFX 96 real-time system thermocycler (BioRad). For each run, laboratory strain of P. falciparum 3D7 and nuclease free water was used as positive and negative controls respectively.
High Resolution Melting (HRM) for P. Falciparum drug genotyping
Evaluation of single nucleotide polymorphism associated with resistance to pyrimethamine Pfdhfr (codons 51, 59, 108,164), sulphadoxine -Pfdhps (codons 436, 437, 540, 581, 613), artemisinin-partner drug such as lumefantrine and amodiaquine - Pfmdr (86, 184, 1042, 1246), and chloroquine Pfcrt (72–76) were done using the Qiagen TypeIT master mix. First, the primers were reconstituted and diluted to 10X, from this, 0.7µM and 1X of Qiagen master mix, 3.3 Μl of nuclease free water and 1 µL of gDNA (Supplementary Table 1) was used for the HRM drug assay as described earlier [31] For each drug target, mutant and wild type strains of laboratory cultured adapted parasites and nuclease free water were used as controls.
Statistical analysis
Data was entered in excel and exported to Statistical Package for Social Sciences (SPSS) version 21.0 (SPSS, Inc. Chicago IL, USA) for analyses. The performance of each diagnostic tool was evaluated as per their sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) with RT- PCR as the gold standard. Agreement between pairs of evaluation tools was tested based on Cohen Kappa’s statistics where ≤ 0 indicates no agreement, 0.01–0.40 - slight to an average agreement, 0.41–0.80– moderate-stable agreement, and 0.81-1.00 perfect agreement [48].
Results of the drug resistant assay were scored using the Light-cycler software supplied with the machine after adjusting the sliding window to the appropriate melt curve. Samples with the same curve profiles with either the mutant or wild type controls were scored accordingly (wild or mutant or mixed in cases where it has the curves of both strain).
Further, amino acid mutation of the single nucleotide polymorphisms at each codon of each target molecular marker (Pfcrt, Pfdhfr, Pfdhps and Pfmdr1) were used in constructing the different haplotype per specific gene.
Results
Participants background information and parasite diagnosis
Majority of the pregnant women sampled was within the age brackets 30–34 (33.1%) and 25–29(33.1%). This was followed by the age brackets 20–24(21.5%) and 35–39(10.7%). Only one participant each falls within the age brackets 15–19 (0.8%) and 40–44 (0.8%). There was more Multigravida − 86(71.1%) than Primigravida– 35(28.9%) women in the study. In terms of pregnancy terms, the highest participants were on their second trimester (51.2%) followed by the third trimester (48.0%), while the least were the first trimester (0.8%). With regards to the use of IPTp with sulphadoxine pyremethamine, surprisingly, majority (59.5%) of them did not receive the preventive drug prior to the day that they were enrolled in the study, and only 24 (19.8%) of them had taken one dose, 18(14.9%) had taken two doses, while only 7(5.8%) of them had used three doses (Table 1 below).
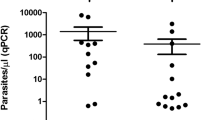
One hundred and twenty-one (52 from Kosofe and 69 from Epe) pregnant women were recruited into this study. Of this, rapid diagnosis with P. falciparum HRP2-mRDT detected malaria infections in eight (0.06%) of them, the ultra-sensitive Alere™ Pf malaria RDT also gave similar outcome in the same samples as detected by the mRDT (Fig. 1). In contrast, microscopy and RT-PCR confirmed only four of the eight infections detected by both rapid diagnostic tests to be true positive and RT-PCR detected additional three samples not shown to be positive by either of the mRDT. Thus, employing RT-PCR as the comparator (gold standard), PfHRP2 and uRDT both showed high specificity (96.9% ), but their sensitivities were low (Table 2). However, the sensitivity and specificity of microscopy with regards to RT-PCR was almost perfect (96.69% and 96.58% respectively). For all further drug genotyping assay, only the seven samples positive by both microscopy and RT-PCR were included.
Drug resistant genotypes of Pfdhps and Pfdhfr from the study participants
Single nucleotide polymorphism in Pfdhps gene associated with sulphadoxine resistant revealed the presence of S613 mutant genotypes in three of the seven positive isolates and isolates with mixed wild/mutant genotype (A613S) at this codon. In addition, four mixed genotypes at the A581G codon was also recorded while the other Pfdhps codons showed the presence of wild type alleles.
In the Pfdhfr gene associated with pyrimethamine resistant, we observed mutations in 28.6%, 28.6%, 85.7% at the I51, R59 and N108 codons respectively. Mixed wild and mutant type genotypes were also observed in 28.6% at each of the N51I, and C59R codons respectively (Fig. 2).
Profiles of malaria resistant markers of Pfmdr1 and pfcrt from the study participants
The P. falciparum multi-drug resistant gene 1 which has been associated with reduced parasite tolerance to amodiaquine and lumefantrine was also genotyped. Of the seven samples assayed, 85.7%, 71.4% and 14.3% harboured parasites that are resistant at the 86Y, 184 F and 1042D codons respectively. All parasites were of the wild type allele for the D1246 codon (Fig. 3). The Pfcrt mutant haplotype (CVIET) was observed in 14.3% of the isolates and a mixed mutant/wild genotype (14.3%) at these codons (72–76) was also observed (Table 3).
Haplotypic frequency and prevalence of the different drug resistant genes
Parasites with the Pfdhps single mutant SAKAS (with mutation at the 613 codon) were the most common occurring in 42.9% of the isolates while the single mutant Pfdhfr NCNI was the most common observed in these pregnant women, followed by the triple mutant IRNL (28.6%). In the Pfmdr1 gene, double mutant (YFND) was the most prevalent (57.1%) while the other single mutants YYND, NFND and YYDD occurred in similar proportion (14.3%). The CVIET triple codon mutation and the triple mutant + wild type has similar proportion (14.3%) (Table 3 above).
Discussion
Malaria in pregnancy remains a public health challenge especially in malaria endemic areas such as Nigeria, and as a result of the growing evolution of malaria drug resistance, effective treatment of pregnant women with malaria is now more than ever threatened. The study evaluated four malaria diagnostic tools among pregnant women suspected of malaria infections and characterized Pfdhfr, Pfdhps, Pfmdr1, and Pfcrt drug resistant markers in malaria positive isolates. One hundred and twenty-one women were enrolled into this study and majority of them were in the age brackets 20–39 years. In addition, majority of the women had had multiple pregnancies before (multigravida), and unfortunately many of the recruited women had not taken IPTp-SP which poses a disturbing scenario, as this will result in the continuous transmission of P. falciparum with detrimental outcome to both the pregnant woman and the foetus. The study showed concordant results for both the PfHRP2 and the ultra-sensitive Alere mRDTs in terms of sensitivity and specificity. This is similar to the findings of Unwin et al., 2020 [19]. Both of them showed high specificity and low sensitivity. In contrary, Briand et al., 2020 [32] reported that the ultra-sensitive rapid diagnostic test - uRDT specificity was slightly lower than that for conventional mRDT among Beninese pregnant women. The sensitivity from their study was high particularly among those in their first trimester, the multigravidae and asymptomatic. In addition, a Colombian study by Vasquez et al., 2018 also demonstrated a non-significant higher sensitivity of uRDT than Standard Bioline (sdRDT) [33]. Differences in transmission dynamics, endemicity and parasite density in these study areas could have been responsible for the variation in the performance of the mRDT tools.
Our microscopy and real-time PCR (RT-PCR) confirmed only four out of the eight infections detected by both mRDTs to be truly positive and further detected three samples also classified as negative to be positive for falciparium malaria. Misclassification of results as false negative by any of these mRDTs has serious implications to maternal and child health on one hand, and continuous malaria transmission on the other hand. The importance of diagnosis cannot be overemphasized as it is a prerequisite for treatment. Therefore, in order not to miss submicroscopic infections in pregnant women and avoid the deleterious effects of PAM, more sensitive tools should be employed. uRDT has been considered to be better in terms of sensitivity and specificity, however varying low sensitive and specificity results are presented from different studies and so there is need for re-evaluation of the efficiency of the uRDT. In addition to this it is very expensive when compared with the conventional mRDT.
Drug resistant typing was carried out on codons A613S, A581G, G436S, G437 and K540E for Pfdhps gene; N51,I C59R S108N, and I164L for Pfdhfr gene; 72–76 for Pfcrt and codons N86Y, Y184F, N1042D and D1246Y for Pfmdr1. Haplotypic distribution and prevalence of the different drug resistant genes were also estimated. Single nucleotide polymorphism (SNP) data from our study showed high prevalence of single mutant N108Y (57.1%), triple mutant N51I, C59R, S108N (28.6%) Pfdhfr alleles and single mutant A613S/T (42.9%) Pfdhps allele.
The single mutant haplotype- SAKAS in Pfdhps was the most prevalent haplotype (42.9%) while single mutation (NCNI) in Pfdhfr also was the most prevalent haplotype (57.1%) from our study. Our finding is dissimilar to that of Lucchi et al., 2015 [34] which noted that prevalence of these haplotypes in West Africa are generally high. Although, the prevalence observed here is low compared to a previous study conducted elsewhere in Nigeria; [26] in Equatorial Guinea [35] and in Democratic Republic of Congo [36]. This could be due to the small number of sample size used for the drug resistant assay in the present study. However, lower prevalence (26.5– 56.25%) has also been reported in other West African countries such as in Senegal [29] and in Bukina Faso [37]. In our study, wild type alleles were recorded for S436, A437 and K540 of Pfdhps and I164 of the Pfdhfr. Mixed wild and mutant alleles were recorded for A581G of the Pfdhps as well. As per the WHO recommendation [38]for the discontinuation of IPT-SP in areas where K540E mutation prevalence is > 95% and A581G > 10% constant surveillance should be taken seriously in the study areas to be able to identify and track any change in the distribution of mutation that would inform policy on the IPT-SP use.
In our study, polygenomic infection was observed in A581G, and A613S/T of the Pfdhps alleles and N51I, C59R and S108N of the Pfdhfr alleles. The mutations in these two (Pfdhfr and Pfdhps) alleles were high. This is in agreement with the findings of Adegbola et al., 2023 [47] which reported a relatively high prevalence of SNPs from both Pfdhps and Pfdhfr genes. They also reported a relatively high prevalence of SNPs from both Pfdhps and Pfdhfr genes. They also reported that sextuple and septuple contributed to about 25.0% of the P. falciparum isolates in their study and opined that their presence might pose a high probability of the malaria parasite becoming extensively resistant to SP in Nigeria.
Quadruple[ [26, 27] (reported earlier in Nigeria), quintuple and sextuple [31, 40, 41](reported in South and East Africa) mutant haplotypes which had been linked with both in vivo and in vitro SP resistance were not observed in our study [39,40,41].
For the Pfmdr1, out of the seven samples assayed, 85.7%, 71.4% and 14.3% harboured parasites with resistant alleles at the 86Y, 184 F and 1042D codons respectively. This is similar to the findings of Issa et al., 2022 and Adamu et al., 2020 [41, 42] where Pfmdr1 184 F and 86Y predominated in their studies in Niger Republic and Northern part of Nigeria respectively. The high prevalence of Pfmdr1 86Y alleles has been associated with chloroquine resistance. This might be an indication of the risk in the efficacy of Arthemeter Lumefanterine (AL). Prevalence of Pfmdr1 N86 allele seen in our study might be suggestive of possible AL pressure in the population. The Pfmdr1 double mutant 86Y/184F was found in 57.1% of our study. This is in line with the findings of Issa et al., 2022 and Tuedom et al., 2021 [42, 43]. While the S1034C and D1246Y mutations were detected in our study, they were not found in the study by Issa et al., 2022 [42]. For the Pfcrt, it has been established that the CVIET haplotype is widely prevalent in Nigeria from various studies [44,45,46]. Our study reported 14.3% CVIET haplotype distribution which is lower than what Issa et al., 2022 reported [42]. They reported a higher 33.07% isolates from their study that harboured the CVIET mutant haplotype.
Conclusion
Although, this study is limited in the number of sample size included in the study and the geographical spread of the samples, however, it emphasize the importance of the use of high sensitive tools for the diagnosis of malaria especially because of the submicroscopic infections that might not be detected with less sensitive tools. Relying solely on the outcome of RDT should be done with caution since misclassification of results as false negative by the mRDTs is possible as evident in this study. This has implication to the maternal, neonatal and child health and ultimately will perpetuate the continuous transmission of malaria. The presence of polygenomic infection which is indicative of high parasite recombination events and the possibility of spread of mutant parasite strains due to high transmission has the potential of jeopardizing the use of SP- IPTP in the study area. Therefore, continuous monitoring should be done so as to identify presence of mutation and take appropriate and prompt action especially considering the fact that SP is the only available preventive treatment for the pregnant women.
Data availability
The datasets generated and analyzed in this study have been included in this manuscript.
Abbreviations
- DNA:
-
Deoxyribonucleic Acid
- HRP-2:
-
Histidine - Rich Protein 2
- IPTp-SP:
-
Intermittent Preventive Treatment in Pregnancy with Sulphadoxine Pyrimethamine
- mRDT:
-
Malaria Rapid Diagnostic Test
- NPV:
-
Negative Predicative Value
- LSH&TM:
-
London School of Hygiene & Tropical Medicine
- MRC:
-
Medical Research Council
- NIMR:
-
Nigerian Institute of Medical Research
- PAM:
-
Pregnancy Associated Malaria
- Pfcrt:
-
Plasmodium falciparum Chloroquine Resistant Transporter
- Pfdhfr:
-
Plasmodium falciparum dihydrofolate reductase
- Pfdhps:
-
Plasmodium falciparum dihydropteroate synthase
- Pfmdr1:
-
Plasmodium falciparum Multidrug Resistance
- PPV:
-
Positive Predicative Value
- RT-PCR:
-
Real Time Polymerase Chain Reaction
- uRDT:
-
Ultra Sensitive Rapid Diagnostic Test
- varATS:
-
var Acid Terminal Segment
- WHO:
-
World Health Organization
References
WHO. 2020. World malaria report 2019. Geneva, World Health Organization, 2019.
Mendez C. Malaria during pregnancy: a priority area of malaria research and control. Parasitol Today. 1995;11:179–83.
Moya-Alvarez V, Abellana R, Cot M. Pregnancy-associated malaria and malaria in infants: an old problem with present consequences. Malar J. 2014;13:271.
World Health Organisation. WHO policy brief for the implementation of intermittent preventive of malaria in pregnancy using sulfadoxin-pyrimethamine (IPTp-SP). WHO Global Malaria Programme; 2014.
Rogerson SJ, Mwapasa V, Meshnick SR. Malaria in pregnancy: linking immunity and pathogenesis to prevention. Am J Trop Med Hyg. 2007;77:14–22.
Le Hesran J-Y, Cot M, Personne P, Fievet N, Dubois B, Beyeme M, Boudin C, Deloron P. Maternal placental infection with Plasmodium Falciparum and malaria morbidity during the first 2 years of life. Am J Epidemiol. 1997;146:826–31.
Brabin B. The risk and severity of Malaria in pregnant women. World Health Organisation; 1991.
Thompson JM, Eick SM, Dailey C, Dale AP, Mehta M, Nair A, Cordero JF, Welton M. Relationship between pregnancy-Associated Malaria and adverse pregnancy outcomes: a systematic review and Meta-analysis. J Trop Pediatr. 2020;66:327–38.
Umbers AJ, Aitken EH, Rogerson SJ. Malaria in pregnancy: small babies, big problem. Trends Parasitol. 2011;27:168–75.
Steketee RW, Nahlen BL, Parise ME, Menendez C. The burden of malaria in pregnancy in malaria-endemic areas. Am J Trop Med Hygiene. 2001;64:28–35.
World Health Organisation. 2015. Guidelines for the Treatment of Malaria, 3rd edition. Geneva, Switzerland:WHO.
Ohrt C, Purnomo M, Sutamihardja MA, Tang D, Kain KC. Impact of microscopy error on estimates of protective efficacy in malaria-prevention trials. J Infect Dis. 2002;186:540–6.
Payne D. Use and limitations of light-microscopy for diagnosing malaria at the primary health-care level. Bulleting World Health Organisation. 1988;66:621–6.
Mathison BA, Pritt BS. Update on malaria diagnostics and test utilization. J Clin Microbiol. 2017;55:2009–17.
Maltha J, Gillet P, Jacobs J. Malaria rapid diagnostic tests in endemic settings. Clin Microbiol Infect. 2013;19:399–407.
Gerstl S, Dunkley S, Mukhtar A, De Smet M, Baker S, Maikere J. Assessment of two malaria rapid diagnostic tests in children under five years of age, with follow-up of false-positive pLDH test results, in a hyperendemic falciparum malaria area, Sierra Leone. Malar J. 2010;9:28.
World Health Organisation. Response plan to Pfhrp2 gene deletions. Geneva: World Health Organisation; 2019.
Bosco AB, Nankabirwa JI, Yeka A, Nsobya S, Gresty K, Anderson K, Mbaka P, Prosser C, Smith D, Opigo J, Namubiru R, Arinaitwe E, Kissa J, Gonahasa S, Won S, Lee B, Lim CS, Karamagi C, Cheng Q, Nakaaga JK, Kama MR. Limitations of rapid diagnostic tests in malaria surveys in areas with varied transmission intensity in Uganda 2017–2019: implications for selection and use of HRP2 RDTs. PLoS ONE. 2020;15(12):e0244457.
Unwin VT, Ahmed R, Noviyanti R, Puspitasari AM, Utami RAS, Trianty L, Lukito T, Syafruddin D, Poespoprodjo JR, Santana-Morales MA, Kuile FOT, Adams ER. Use of a highly–sensitive rapid diagnostic test to screen for malaria in pregnancy in Indonesia. Malar J. 2020;19:28.
Acquah FK, Donu D, Obboh EK, Bredu D, Mawuli B, Amponsah JA, Quarte J, Amoah LE. Diagnostic performance of an ultrasensitive HRP2–based malaria rapid diagnostic test kit used in surveys of afebrile people living in Southern Ghana. Malar J. 2021;20:125.
Kayentao K, Garner P, van Eijk AM. Intermittent preventive therapy for malaria during pregnancy using 2 vs 3 or more doses of sulfadoxine-pyrimethamine and risk of low birth weight in Africa: systematic review and meta-analysis. JAMA. 2013;309:594–604.
Oyibo WA, Agomo CO. Scaling up of intermittent preventive treatment of malaria in pregnancy using sulphadoxine-pyrimethamine: prospects and challenges. Maternal Child Health J. 2011;15:542e552.
Menéndez C, Bardají A, Sigauque B, Sanz S, Aponte JJ, Mabunda S. (2010) Malaria prevention with IPTp during pregnancy reduces neonatal mortality. PLoS ONE. 5:e9438. practices among mothers delivering in an urban hospital in south west Nigeria. Journal of Vector Borne Diseases 45:217– 24.
Happi CT, Gbotosho GO, Folarin OA, Akinboye DO, Yusuf BO, Ebong OO. Polymorphisms in Plasmodium Falciparum dhfr and dhps genes and age related in vivo sulfadoxine–pyrimethamine resistance in malariainfected patients from Nigeria. Acta Trop. 2005;95:183–93.
Mockenhaupt FP, Teun Bousema J, Eggelte TA, Schreiber J, Ehrhardt S, Wassilew N, Otchwemah RN, Sauerwein RW, Bienzle U. Plasmodium Falciparum dhfr but not dhps mutations associated with sulphadoxine-pyrimethamine treatment failure and gametocyte carriage in northern Ghana. Trop Med Int Health. 2005;10(9):901–8.
Kayode AT, Ajogbasile FV, Akano K, Uwanibe JN, Oluniyi PE, Eromon PJ, Folarin OA, Sowunmi A, Wirth DF, Happi CT. Polymorphisms in Plasmodium Falciparum dihydropteroate synthetase and dihydrofolate reductase genes in Nigerian children with uncomplicated malaria using high–resolution melting technique. Nat Res. 2021;11:471.
Oguike MC, Falade CO, Shu E, Enato IG, Watila I, Baba ES, Bruce J, Webster J, Hamade P, Meek S, Chandramohan D, Sutherland CJ, Warhurst D, Roper C. Molecular determinants of sulfadoxine-pyrimethamine resistance in Plasmodium Falciparum in Nigeria and the regional emergence of dhps 431V. Int J Parasitology: Drugs Drug Resist. 2016;6:220–9.
Okwa OO, Akinmolayan FI, Carter V, Hurd H. Transmission dynamics of malaria in four selected ecological zones of Nigeria in the rainy season. Ann Afr Med. 2009;8(1):1–9.
Hofmann N, Mwingira F, Shekalaghe S, Robinson LJ, Mueller I, Felger I. Ultra-sensitive detection of Plasmodium falciparum by amplification of Multi-copy Subtelomeric targets. PLoS Med. 2015;12(3):e1001788.
Ndiaye YD, Diedhiou CK, Bei AK, Dieye B, Mbaye A, Mze NP, Daniels RF, Ndiaye IM, Deme AB, Gaye A, Sy M, Ndiaye T, Badiane AS, Ndiaye M, Premji Z, Wirth DF, Mboup S, Krogstad D, Volkman SK, Ahouidi AD, Ndiaye D. High-resolution melting: a useful field-deployable method to measure dhfr and dhps drug resistance in both highly and lowly endemic plasmodium populations. Malar J. 2017;16:153.
Daniels R, Ndaye D, Wall M, McKinney J, Sene PD, Sabeti PC, Volkman SK, Mboup S, Wirth DF. Rapid, Field-Deployable Method for genotyping and Discovery of single-nucleotide polymorphisms Associated with Drug Resistance in Plasmodium Falciparum. Antimicrob Agents Chemother. 2012;56(6):2976–86.
Briand V, Cottrell G, Ndam NT, Vendrell XM, Vianou B, Mama A, Kouwaye B, Houzé S, Bailly J, Gbaguidi E, Sossou D, Massougbodji A, Accrombessi M, Mayor A, Ding XC, Fievet N. Prevalence and clinical impact of malaria infections detected with a highly sensitive HRP2 rapid diagnostic test in Beninese pregnant women. Malar J. 2020;19:188.
Vasquez AM, Medina AC, Tobon-Castano A, Posada M, Velez GJ, Campillo A, Gonzalez IJ, Ding X. Performance of a highly sensitive rapid diagnostic test (HS-RDT) for detecting malaria in peripheral and placental blood samples from pregnant women in Colombia. PLoS ONE. 2018;13(8):e0201769.
Lucchi NW, Okoth SA, Komino F, Onyona P, Goldman IF, Ljolje D, Shi YP, Barnwell JW, Udhayakumar V, Kariuki S. (2015). Increasing prevalence of a novel triple-mutant dihydropteroate synthase genotype in Plasmodium falciparum in western Kenya. Antimicrob. Agents Chemother 59(7), 3995–4002.Maltha J, Gillet P and Jacobs J.(2013) Malaria rapid diagnostic tests in endemic settings. Clinical Microbiology & Infection19:399–407.
Jiang T, Chen J, Fu H, Wu K, Yao Y, Eyi JUM, Matesa RA, Obono MMO, Du W, Tan H, Lin M, Li J. High prevalence of pfdhfr–pfdhps quadruple mutations associated with sulfadoxine–pyrimethamine resistance in Plasmodium Falciparum isolates from Bioko Island, Equatorial Guinea. Malar J. 2019;18:101.
Mobula L, Lilley B, Tshefu AK, Rosenthal PJ. Resistance-mediating polymorphisms in Plasmodium falciparum infections in Kinshasa, Democratic Republic of the Congo. Am J Trop Med Hygiene. 2009;80:555–8.
Tahita MC, Tinto H, Erhart A, Kazienga A, Fitzehenry R, VanOvermeir C, Rosanas-Urgell, Ouedraogo J, Guiguemde RT, Vangeertruyden JP, D’Alessandro U. Prevalence of the dhfr and dhps mutations among pregnant women in rural Burkina Faso five years after the introduction of intermittent preventive treatment with sulfadoxine-pyrimethamine. PLoS ONE. 2015;10(9):e0137440–0137440.
World Health Organisation. WHO Policy brief for the implementation of intermittent preventive treatment of malaria in pregnancy using sulfadoxine-pyrimethamine (IPTp-SP). Geneva: World Health Organization; 2013.
Naidoo I, Roper C. Drug resistance maps to guide intermittent preventive treatment of malaria in African infants. Parasitology. 2011;138:1469–79.
Bwijo B, Kaneko A, Takechi M, Zungu IL, Moriyama Y, Lum K, Tsukahara T, Mita T, Takahashi N, Berggvist Y, Bjorkman A, Kobayakawa T. High prevalence of quintuple mutant dhps/dhfr genes in Plasmodium falciparum infections seven years after introduction of sulfadoxine and pyrimethamine as first line treatment in Malawi. Acta Trop. 2003;85:363–73.
Roper C, Pearce R, Nair S, Nosten F, Anderson T. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124.
Issa I, Lamine MM, Hubert V, Ilagouma A, Adehossi E, Mahamadou A, Loobo NF, Sarr D, Shollenberger LM, Sandrine H, Jambou R, Laminou IM. Prevalence of mutations in the Pfdhfr, Pfdhps, and Pfmdr1 genes of Malarial parasites isolated from symptomatic patients in Dogondoutchi, Niger. Trop Med Infect Dis. 2022;7:155.
Tuedom AGB, Sarah-Matio EM, Moukoko CEE, Feufack-Donfack BL, Maffo CN, Bayibeki AN, Awono-Ambene HP, Ayong L, Berry A, Abate L, Morlais I, Nsango SE. Antimalarial drug resistance in the Central and Adamawa regions of Cameroon: prevalence of mutations in P. Falciparum Crt, Pfmdr1, Pfdhfr and pfdhps genes. PLoS ONE. 2021;16(8):e0256343. https://doi.org/10.1371/journal.
Adams R, Mukhtar MM, Abubakar UF, Damudi HA, Muhammad A, Ibrahim SS. Polymorphism analysis of Pfmdr1 and Pfcrt from Plasmodium Falciparum isolates in Northwestern Nigeria revealed the Major Markers Associated with Antimalarial Resistance. Diseases. 2021;9:6.
Gbotosho GO, Folarin OA, Bustamante C, Pereira da Silva LH, Mesquita E, Sowunmi A, Zalis MG, Oduola AMJ, Happi CT. Different patterns of Pfcrt and Pfmdr1 polymorphisms in P. Falciparum isolates from Nigeria and Brazil: the potential role of Antimalarial Drug Selection pressure. Am J Trop Med Hygiene. 2012;88(2):211–3.
Ikegbulam MN, Nkonganyi CN, Thomas AN, Esimone CO, Velavan TP, Ojurongbe O. Analysis of Plasmodium Falciparum Pfcrt and Pfmdr1 genes in parasite isolates from asymptomatic individuals in Southeast Nigeria 11 years after withdrawal of chloroquine. Malar J. 2019;18:343.
Adegbola AJ, Ijarotimi OA, Ubom AE, Adesoji BA, Babalola OE, Hocke EF, Hansson H, Mousa A, Bolaji OO, Alifrangis M, Roper C. A snapshot of the prevalence of dihydropteroate synthase-431V mutation and other sulfadoxine-pyrimethamine resistance markers in Plasmodium Falciparum isolates in Nigeria. Malar J. 2023;22:71.
McHugh ML. Interrater reliability: the kappa statistic. Biochemia Med. 2012;22(3):276–82.
Acknowledgements
We thank all the pregnant participants for their willingness to enroll in the study. We thank the Pharmanews and Emzor Pharmaceuticals for providing Sulfadoxine-Pyrimethamine (Maldox) for the participating health facilities for pregnant women. We also thank the Director, Medical Services and Disease control, Lagos State Primary Health care board, the laboratory scientists and the Medical Officers of health at Ogudu and Epe Primary Health Centres.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not for profit sectors. The research work was self-sponsored.
Author information
Authors and Affiliations
Contributions
A. I and M. O carried out the study design and manuscript writing. A. I collected blood samples. M.O. did data analysis. F.F and E.A. carried out laboratory experiment. O.A., E.I and A, N reviewed and critiqued the manuscript.
Corresponding author
Ethics declarations
Ethical approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki, and the ethical approval (IRB/17/048) was obtained from the ethical review board of the Nigerian Institute of Medical Research (NIMR), Yaba, Lagos. Verbal and written informed consent was obtained from all pregnant women included in this study after detailed explanation of the study objectives to each of them. Pregnant women included in this study are those that consented with no pregnancy-related complications.
Consent for publication
All the authors have given consent for the publication of this work.
Consent for publication from participants
Not applicable.
Limitations of study
This study is limited in the number of sample size included in the study and the geographical spread of the samples because it was self-funded.
Competing interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Ibekpobaoku, A.N., Oboh, M.A., Faal, F. et al. Sub-microscopic Plasmodium falciparum infections and multiple drug resistant single nucleotide polymorphic alleles in pregnant women from southwestern Nigeria. BMC Res Notes 17, 129 (2024). https://doi.org/10.1186/s13104-024-06763-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-024-06763-2