Abstract
Objectives
Citrus yellow vein clearing virus (CYVCV) is an emerging disease that poses a significant threat to the citrus industry in California. In this study, the viral genomic RNA was isolated from Eureka lemon plants in the greenhouse exhibiting CYVCV symptoms. Subsequently, the corresponding DNA genome amplicon was sequenced and annotated. These efforts expand the genotype database of CYVCV, which aims to enhance detection assays, promote understanding of the virus’s genetics and evolution, and support the management of this disease.
Data description
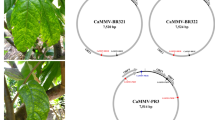
In this report, we present the complete genome sequence of the CYVCV California isolate (CA1). The genome was found to be 7,530 bp in length, with a G + C content of 51.7%. The 5′ and 3′ termini were determined using 5′ and 3′ termini rapid amplification of cDNA ends (RACE) systems. Furthermore, our analysis revealed the presence of six open reading frames (ORFs) potentially encoding proteins. All sequence data and annotation have been deposited in GenBank under the accession number OR037276.1.
Similar content being viewed by others
Objective
Citrus yellow vein clearing virus (CYVCV) is a serious emerging quarantine disease that poses a threat to the citrus industry in California and across the United States. The symptoms of yellow vein clearing disease induced by the virus vary depending on citrus varieties and environmental conditions [1, 2]. Lemon (Citrus limon) and sour orange (C. aurantium) trees are severely affected, while a wide range of other citrus cultivars are susceptible but remain asymptomatic [1, 3]. Infected citrus trees displaying symptoms suffer from growth deficiencies, reduced citrus production, accompanied by yellow vein clearing, adaxial water-soaked appearance of veins, leaf distortion, occasional ringspots, and venial necrosis [1,2,3].
CYVCV can be transmitted through grafting and mechanical means and is naturally vectored by aphids and whiteflies [1, 4]. The first occurrence of yellow vein clearing disease in citrus was reported in lemons and sour orange from Pakistan in 1988 [5]. Since then, it has been detected in Turkey, India, Iran, and China. The rapid spread of CYVCV in China since 2009 has resulted in serious losses in lemon production [6, 7]. In 2022, the routine multi-pest survey conducted by the California Department of Food and Agriculture (CDFA) identified CYVCV-infected citrus trees in urban properties in the cities of Tulare and Visalia, while no CYVCV have been detected in commercial citrus orchards. The United States Department of Agriculture, Animal and Plant Health Inspection Service (USDA, APHIS) has confirmed positive identifications of the disease. The CDFA currently rates CYVCV as a pest of high concern (Pest rating A) [8].
CYVCV belongs to the Alphaflexiviridae virus family, Mandaraivirus genus, and is a positive-sense flexuous RNA virus [9]. To investigate potential genomic variations among CYVCV populations in California and other geographical regions, the complete genome sequence of the CYVCV CA1 isolate was amplified and sequenced using state-of-the-art long-read sequencing technology (Plasmidsaurus, Oregon). The assembled complete genome sequence was annotated and deposited in GenBank under the accession number OR037276.1. These genome sequences will be utilized for comparative genomics, contributing to a better understanding of the etiology, relationships, and evolution of CYVCV. Moreover, this genomic data will enhance the efficiency of detection assays for CYVCV in the United States.
Data description
The citrus buds containing CYVCV isolates was collected from a lemon tree in Tulare, California, and grafted onto Eureka lemon seedlings. The grafted plants were maintained in an air-conditioned quarantine greenhouse at the San Joaquin Valley Agricultural Sciences Center in Parlier, California. Total RNA was isolated from a lemon leaf exhibiting yellow vein clearing symptoms using Trizol Reagent (Thermofisher Scientific, USA). A conserved region in the 5′ end, identified through alignment with other reported CYVCV isolates, was utilized to design a virus-specific 5’ race primer (5’-GGTTAGTGGTATTGCCCTGTT-3’). An oligo(dT) primer was used as the 3’ race-specific primer. The 5’ race and 3’ race polymerase chain reaction (PCR) amplicons were purified and cloned into the pGEM-T easy vector (Promega Corp., Wisconsin). The constructed vectors were further sequenced (Plasmidsaurus, Oregon) to obtain the sequences of the CYVCV 5′ and 3′ termini.
Using the 5′ and 3′ termini sequences, the full genome sequence of the CYVCV CA1 isolate was amplified using the Q5 high-fidelity enzyme (New England Biolabs Inc. Ipswich, Maryland) and virus-specific PCR primers (5′ primer-GAAAAGCAAACATAACCAACACACACCC; 3′ primer-CAGAAAATGGAAACTGAAAGCCTGAATATTT). The resulting 7.5 Kb PCR amplicon was sequenced using the latest long-read sequencing technology from Oxford Nanopore Technologies (ONT, Plasmidsaurus, Oregon). The sequencing process involved constructing an amplification-free long-read sequencing library using the newest v14 library prep chemistry, minimal fragmentation of the linear input DNA in a sequence-independent manner, primer-free sequencing of the library using the highly accurate R10.4.1 flow cells (raw data is > 99% accurate), and re-assembly of the raw reads by aligning them against each other to generate a high-accuracy linear consensus sequence (Data file 1, Table 1).
The genome of the CYVCV CA1 isolate was found to be 7,530 bp in length, with an average G + C content of 51.7%. The genome sequence data has been deposited in GenBank under the accession number OR037276.1. The six Open Reading Frames (ORFs) were annotated as follows: ORF1 ranges from 80 bp to 5,029 bp; ORF2 ranges from 5,036 bp to 5,714 bp; ORF3 ranges from 5,691 bp to 6,017 bp; ORF4 ranges from 5,944 bp to 6,126 bp; ORF5 ranges from 6,149 bp to 7,126 bp; ORF6 ranges from 6,826 bp to 7,494 bp. The 5′ and 3′ untranslated regions (UTRs) were also characterized. These data significantly expand the sequence database of CYVCV and are expected to enhance detection assays and provide insights into the evolution of this plant pathogenic virus.
Limitations
The CYVCV CA1 isolate was collected from a single lemon plant.
Data Availability
The data described in this Data note can be freely and openly accessed on Genbank under accession number OR037276.1. Please see Table 1 and references [10] for details and links to the data.
Abbreviations
- BP:
-
Base pair
- CDFA:
-
California Department of Food and Agriculture
- CYVCV:
-
Citrus yellow vein clearing virus
- ORF:
-
Open Reading Frame
- RACE:
-
Rapid amplification of cDNA ends
- PCR:
-
Polymerase chain reaction
- USDA, APHIS:
-
United States Department of Agriculture, Animal and Plant Health Inspection Service
- UTR:
-
Untranslated region
References
Liu C, Liu H, Hurst J, Timko MP, Zhou C. Recent advances on Citrus yellow vein clearing virus in Citrus. Hortic. Plant J. 2020;6(4):216–22.
Chen HM, Li ZA, Wang XF, Zhou Y, Tang KZ, Zhou CY, Zhao XY. First report of Citrus yellow vein clearing virus on lemon in Yunnan, China. Plant Dis. 2014;98(12):1747.
Bin Y, Xu J, Duan Y, Ma Z, Zhang Q, Wang C, Su Y, Jiang Q, Song Z, Zhou C. The Titer of Citrus Yellow Vein Clearing Virus is positively Associated with the severity of symptoms in infected Citrus seedlings. Plant Dis. 2022;106(3):828–34.
Zhang YH, Wang YL, Wang Q, Cao MJ, Zhou CY, Zhou Y. Identification of Aphis spiraecola as a vector of Citrus yellow vein clearing virus. Eur J Plant Pathol. 2018;152:841–44.
Catara A, Azzaro A, Davino M, Polizzi G. Yellow vein clearing of lemon in Pakistan. Proceedings of the 12th Conference of the International Organization of Citrus Virologists, Riverside, CA. 1993. 364 – 67.
Chen HM, Zhou Y, Wang XF, Zhou CY, Yang XY, Li ZG. Detection of Citrus yellow vein clearing virus by quantitative real-time RT-PCR. Hortic. Plant J. 2016;2(4):188–92.
Liu YJ, Wang YL, Wang Q, Zhang YH, Shen WX, Li RH, Cao MJ, Chen L, Li X, Zhou CY, Zhou Y. Development of a sensitive and reliable reverse transcription droplet digital PCR assay for the detection of citrus yellow vein clearing virus. Arch Virol. 2019;164(3):691–97.
Scheck HJ. California pest rating proposal for Citrus yellow vein clearing virus. 2021. https://blogs.cdfa.ca.gov/Section3162/wp-content/uploads/2021/09/Citrus-yellow-vein-clearing-virus-_ADA_PRP-1.pdf.
Loconsole G, Onelge N, Potere O, Giampetruzzi A, Bozan O, Satar S, De Stradis A, Savino V, Yokomi RK, Saponari M. Identification and characterization of citrus yellow vein clearing virus, a putative new member of the genus Mandarivirus. Phytopathology. 2012;102(12):1168–75.
Sun Y, Yokomi RK, GenBank. 2023. https://identifiers.org/nucleotide:OR037276.1.
Acknowledgements
The authors gratefully acknowledge the assistance of Sydney Helm Rodriguez, Biological Science Technician, USDA, ARS, SJVASC, Crop Disease, Pests and Genetics Research Unit, Parlier, CA for the maintenance of the laboratory and plants in the greenhouse. We are also grateful for the support from USDA-APHIS-PPQ-S&T, Raleigh, NC and the CDFA, Sacramento, CA.
Funding
Funds for this research were supported by an USDA, APHIS, PPQ Inter-Agency Agreement (60-2034-3-0002) to RKY. We also acknowledge support from the USDA, ARS, CDPG Project Number 2034-22000-015-000D. The funding bodies played no role in the design of the study and collection, analysis, or interpretation of data or in writing of the manuscript.
Author information
Authors and Affiliations
Contributions
YDS and RKY were involved in conceptualization of the study, analyzing data, and writing the manuscript. RKY acquired and maintained the CYVCV-infected plants. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Sun, Y., Yokomi, R. Whole genome sequence of Citrus yellow vein clearing virus CA1 isolate. BMC Res Notes 16, 166 (2023). https://doi.org/10.1186/s13104-023-06443-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13104-023-06443-7