Abstract
Background
Ticks carry microbes, some of which are pathogenic for humans and animals. To assess this One Health challenge, 342 ticks were collected from pet dogs and cats at 10 veterinary clinics in Finland as part of the European project “Protect Our Future Too”.
Methods
The tick species were identified, and ticks were screened with quantitative PCR (qPCR) for tick-borne pathogens, including Borrelia burgdorferi sensu lato, Borrelia miyamotoi, Ehrlichia canis, Anaplasma spp., Candidatus Neoehrlichia mikurensis, tick-borne encephalitis virus (TBEV), and Babesia spp. For comparison, a subset of tick DNA (20 qPCR-positive samples) was analysed with 16S next-generation sequencing (NGS).
Results
Most ticks were Ixodes ricinus (289, 84.5%), followed by Ixodes persulcatus (51, 14.9%). One hybrid tick (I. ricinus/I. persulcatus, 0.3%) and one Rhipicephalus sanguineus tick (0.3%) were identified. We found one or more of the analysed pathogens in 17% (59/342) of the ticks. The most prevalent pathogen was B. burgdorferi s.l. (36, 10.5%), followed by Anaplasma phagocytophilum (12, 3.5%), B. miyamotoi (5, 1.5%), Babesia venatorum (4, 1.2%), and TBEV (1, 0.3%). Candidatus Neoehrlichia mikurensis DNA was amplified from three (0.9%) ticks. Ehrlichia canis was not detected. In the 16S NGS, six samples produced enough reads for the analysis. In these six samples, we confirmed all the positive qPCR findings of Borrelia spp. and Ca. N. mikurensis.
Conclusions
The high prevalence of pathogenic microorganisms in the ticks of this study emphasizes the importance of awareness of ticks and tick-borne diseases and prevention. Furthermore, the results show that veterinary surveillance can facilitate early detection of tick-borne pathogens and new tick species and draw attention to possible co-infections that should be considered both in symptomatic humans and animals after tick bites.
Graphical Abstract

Similar content being viewed by others
Background
Dogs and cats are the most common pets, and exposure to ticks is inevitable when these animals engage in outdoor activities. Ticks ectoparasitize their hosts, including pets, at each stage of their life cycle and may transmit severe bacterial, parasitic, and viral pathogens. These pathogens can induce behavioural changes in ticks and affect their phenotypic characteristics, making them more active in host-seeking and more resistant against extreme environmental events (e.g., desiccation and cold), which leads to increased fitness and survival [1]. The transmission of pathogens from ticks to hosts depends mainly on the duration of host attachment. For instance, tick-borne encephalitis virus (TBEV) can be transmitted within 60 min of attachment. However, other microorganisms require a longer period [2]. Transmission times may vary considerably between different tick vectors, pathogens, pathogen quantity, host species, and conditions; successful pathogen transmission may require feeding longer than 48 h [3, 4].
Tick-borne infections represent a One Health concern. Without proper protection, tick bites may lead to transmission of tick-borne microorganisms to the animals or the pet owners, which may cause serious and even life-threatening illness. In the northern part of Europe, the most common tick-borne pathogens of zoonotic potential are Borrelia spp., Anaplasma phagocytophilum, and TBEV. However, Rickettsia spp., Candidatus Neoehrlichia mikurensis, and Babesia spp. are also reported [5, 6]. Further, polymicrobial infections can occur, as ticks can carry multiple pathogens simultaneously. This can complicate the diagnosis and management of some cases, especially due to the lack of broad-spectrum diagnostic tools for routine testing and the limited therapeutic options [5, 6].
In Finland, tick-borne infections are mainly transmitted by Ixodes ricinus (dominant in southern Finland) and Ixodes persulcatus (dominant mostly in northern Finland) ticks [7, 8]. Distribution of these ticks is determined by climatic and environmental conditions and animal hosts. In addition, migratory birds and relocated dogs occasionally carry new and rare tick species, which may become endemic in the future climate [9, 10]. According to the Finnish Institute for Health and Welfare, most of the diagnosed and registered human tick-borne infections are Lyme borreliosis (LB; approximately 2000–2500 cases annually) and tick-borne encephalitis (TBE; 151 cases in 2021) [11]. In contrast, a seroprevalence study of Finnish pet dogs suggested that A. phagocytophilum (5.3%) is the most common tick-borne pathogen that dogs encounter, followed by Borrelia burgdorferi (2.9%) and Ehrlichia canis (0.3%) [12]. Another study showed a TBEV seroprevalence of 40% in the Åland Islands and 6% on the south-western archipelago of Finland in samples collected from dogs, which indicates that exposure varies according to geographical location of the animals [13]. This is consistent with the tick abundance and infection prevalence of ticks in a given area.
Several European countries have reported an increase of tick-borne infections, occurring mainly due to changes in climatic and environmental conditions, host reservoir densities, and exposure of human populations [14]. In this context, there is a crucial need for understanding the circulation and the diversity of tick-borne pathogens causing diseases in animals and humans. Pets, particularly dogs, can be used as sentinels for tick-borne diseases. Tick surveillance and analysis in pets can help in estimating the potential risk of these often zoonotic pathogens [15].
As part of the European project “Protect Our Future Too”, which is focused on studying the effects of climate change on pets [16], we performed a study on tick-borne pathogens of veterinary and medical importance in Finland.
Methods
Tick collection
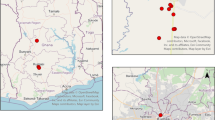
Ticks were collected from pet dogs and cats at 10 veterinary clinics across Finland (Fiskars, Kotka, Turku, Lappeenranta, Pori, Pirkkala, Mikkeli, Jyväskylä, Oulu, and Keminmaa; Fig. 1). Tick collections were performed over a period of 12–16 months by each clinic between June 2020 and November 2021 (Fig. 1). Ticks were removed with Safeguard cards (one for each pet), transferred by pinching the tick legs with tweezers into a sterile tube, and stored refrigerated in RNAlater (Thermo Scientific) until they were shipped to the University of Helsinki for species identification and pathogen screening. Data were recorded upon arrival of a subset of ticks and included information on animal species from which ticks were removed, locality, traveling history of the pet, whether ticks were attached, level of engorgement, developmental status, and sex of ticks.
DNA and RNA extraction
Ticks were homogenized on 96-well plates or in 1.5-ml tubes depending on the number of samples prepared. For homogenization, the ticks were placed individually in the tubes or wells with a 3-mm metal bead and sterile sand. A volume of 400 µl of Dulbecco's phosphate-buffered saline solution with 0.2% bovine serum albumin was added into each tube. Ticks were then homogenized with Qiagen TissueLyser II at a frequency of 30 Hz for 3 min. RNA extraction was performed using a QIAamp Viral RNA Kit (Qiagen) with either the spin protocol (small sample sizes) or the QIAcube (larger sample sizes). DNA extraction was performed on the remaining homogenates with a GeneJET Genomic DNA Purification Kit (Thermo Scientific).
Tick species identification
Identification of tick species was performed primarily by species-specific probes in duplex polymerase chain reaction (PCR) targeting the internal transcribed spacer 2 (ITS2) gene as previously described (Table 1) [17] and by sequencing the same gene [18]. Ticks that were morphologically identified as something other than Ixodes spp. were investigated with PCR and sequencing targeting the ITS2 [18] and cytochrome c oxidase subunit 1 (COX1) genes [19, 20]. The species were verified based on the sequences using the BLAST program available at the National Center of Bioinformatics (NCBI).
Molecular screening of selected pathogens
We used 2× Maxima Probe qPCR Master Mix (Thermo Fischer Scientific) according to the manufacturer’s instructions for DNA detection of B. burgdorferi s.l., B. miyamotoi, Anaplasma spp., E. canis, Ca. N. mikurensis, and Babesia spp. Details on the primers and probes used are described in Table 1. Detection of TBEV RNA was performed with one-step reverse transcriptase PCR (RT-PCR) using TaqMan Fast Virus 1-Step Master Mix (Thermo Fischer Scientific) as described previously [21]. All the reactions included positive and negative controls.
Identification of Anaplasma and Babesia species
A differential qPCR [22] and 2× Maxima Probe qPCR Master Mix were used for identification of A. phagocytophilum. For identification of Babesia species, a traditional PCR protocol [23] using Q5 High-Fidelity PCR Master Mix (New England Biolabs, Inc.) was used. The Babesia PCR products were purified with a GeneJET PCR Purification Kit (Thermo Fisher) and sent to the Institute for Molecular Medicine Finland for Sanger sequencing. The Babesia species were identified using the BLAST program available at the NCBI.
16S next-generation sequencing and analysis
For comparison, we analysed a subset of ticks (n = 20) in which pathogens were detected with specific molecular methods, also with a broad-spectrum method, i.e., 16S next-generation sequencing (NGS). To create libraries, the V3–V4 hypervariable region of the prokaryotic 16S RNA sequence was amplified from the DNA extracts as in the 16S Metagenomic Sequencing Library Preparation guide. Sequencing was performed with an Illumina MiSeq instrument using a MiSeq reagent kit v3. The high-quality reads from each sample were analysed using MG-Rast in R [24] together with in-house scripts for taxonomic profiling of the metagenomic data against the Ribosomal Database Project (RDP) database [25].
Mapping
Occurrence maps of pathogens and tick species were created in Esri ArcGIS (version 10.3.1) (Esri, Redlands, CA, USA). Country boundary shapefiles were downloaded from the open-source spatial database (GADM, 2022).
Results
Ticks and tick species
A total of 342 ticks were collected by veterinary clinics and submitted to the University of Helsinki. Ticks were collected from 30 cats and 259 dogs; host species data were missing for eight animals. Most pets were infested by one tick, but some pets had two to four, and one dog even 15 ticks. Of the 303 ticks from which data were available, 224 (73.9%) were attached, including 118 (38.9% of all) engorged ticks. Seventy-nine (26.1%) were crawling on the animal. Life stage was recorded for 206 ticks and included three nymphs (1.5%), 46 adult males (22.3%), and 157 adult females (76.2%).
The tick species was confirmable by both ITS2 qPCR and ITS2 PCR-based sequencing for 271 ticks. Seventy ticks were identified by ITS2 qPCR only, as we were unable to sequence them despite several attempts. One imported tick from Malaga, Spain, was morphologically identified as not being Ixodes spp., which was confirmed by sequencing the ITS2 and COX1 genes.
Figure 2a shows the location and species of ticks collected by the clinics. The molecular identification of tick species revealed that I. ricinus (289, 84.5%) was predominant in this collection, followed by I. persulcatus (51, 14.9%). One tick (0.3%) showed a hybrid pattern between both species in the duplex qPCR results of the ITS2 gene, showing two amplification plots for both (I. ricinus/I. persulcatus), which indicates that the specific probes for the two species were amplified simultaneously. The tick imported from Spain was identified as Rhipicephalus sanguineus (0.3%) both morphologically and based on sequencing.
Ticks collected from southern, south-western, western, and eastern Finland were identified as I. ricinus. Both I. ricinus and I. persulcatus were collected from pets in central Finland, which is considered a sympatric area for the two tick species. The northernmost tick samples were collected from Southern Lapland (65.8°N) and were identified as I. persulcatus. The ticks were collected over 12 months per clinic, starting from summer of 2020 and ending before the winter of 2021. The earliest ticks in spring were collected on 19 April, and the latest on 24 November.
Pathogen screening
Details and geographical spread of all pathogens detected in ticks in this study are presented in Table 2 and in Fig. 2b–c. A total of 59 of 342 ticks (17.2%) harboured at least one pathogen.
The most prevalent microbe was B. burgdorferi s.l. (36, 10.5%), followed by Anaplasma spp. (12, 3.5%), B. miyamotoi (5, 1.5%), Babesia spp. (4, 1.2%), Ca. N. mikurensis (3, 0.9%), and TBEV (1, 0.3%). Ehrlichia canis was not detected in this collection. All Anaplasma spp. findings were further identified as A. phagocytophilum. Babesia spp. were confirmed as Babesia venatorum. The infection rate differed between the tick species; 56/289 (19.4%) of the I. ricinus ticks and 2/51 (3.9%) of I. persulcatus were infected. The single R. sanguineus was infected with B. venatorum (1/1, 100%). The hybrid tick did not show positive results for the tested pathogens (0/1, 0.0%).
Interestingly, the ticks collected from the dog infested by 15 I. ricinus ticks carried both B. burgdorferi s.l. (four ticks) and A. phagocytophilum (three ticks).
16S NGS data
To assess the potential of NGS for bacterial detection from tick samples, we used 16S sequencing to detect our specific target pathogens (Anaplasma, Borrelia, and Ca. N. mikurensis) in 20 samples. Only six of these samples produced enough reads for the analysis. The bacterial hits against the sequences in the RDP varied from about 8000 to 130 000 (Additional file 1: Table S1). In these six samples, we detected and confirmed the Borrelia and Ca. N. mikurensis qPCR findings. In addition, we detected one Borrelia-positive sample out of these six samples that was negative in the qPCR. We also confirmed that these six samples with enough reads were Anaplasma-negative, confirming the PCR results. Thus, the NGS approach has high potential once optimized for tick samples.
Discussion
Ticks require blood meals to develop to the next life stage. Consequently, infection rate may vary depending on developmental stage. In I. ricinus ticks in Denmark, the infection rate was 2.7 times higher in adults compared to nymphs. Co-infection rates were 12.3% in adult females and 3.5% in nymphs [26]. In our study, 98.5% of the ticks from which data were available were adults. Only 1.5% were nymphs, as these are difficult to detect from pets due to the small size of nymphs and the fur coat of pets, where ticks can easily hide. For the subset of ticks analysed, 38.9% of the ticks were engorged, which, together with the infection rate of 17.2% of these ticks, indicates that these pets were at risk of getting infection. This emphasizes the need for anti-tick medication for pets.
The estimates of tick abundance, microbial content, and infection rate of ticks may differ depending on the collection method [27]. For instance, the commonly used cloth dragging method severely underestimates the abundance of ticks [28]. In Spain, Del Cerro et al. found mostly Borrelia spp. and Rickettsia slovaca in questing ticks only, while some pathogens, including “Candidatus Rickettsia rioja”, Rickettsia raoultii, and A. phagocytophilum were found in both questing ticks and ticks feeding on animals [27]. Additionally, protozoan pathogens were detected in engorged animal-fed ticks except for Babesia bigemina, which was found only in questing ticks collected by dragging [27]. Likewise, in Germany, Babesia spp. and A. phagocytophilum were most prevalent in engorged ticks collected from roe deer, followed by nymphs and adult questing ticks [29].
Our findings agree with previous studies in Finland, which reported I. ricinus and I. persulcatus as the prevalent tick species with medical/veterinary importance and their geographical distribution [7, 8, 30]. These two tick species hybridize naturally, as shown by molecular genetic studies [31]. Here, we found one hybrid tick that did not show a positive result for the studied pathogens. We also found one R. sanguineus that was positive for B. venatorum. It is noteworthy that the dog from which the tick was collected had a travel history with his owner in Spain, explaining its presence in Finland. The risk of importing exotic tick species and pathogens can be increased via traveling with animals without ectoparasitic treatment [10, 32].
Climate and environment play significant roles in the distribution of ticks and tick-borne diseases, as arthropods are especially sensitive to changes in climatic and environmental conditions. Based on the latest climate projections for Finland, mean air temperature is predicted to increase by 2.4 °C in summer and by 3.3 °C in winter by 2070 [33]. Similarly, precipitation is estimated to increase by 5% during summer and by 12% during winter [33]. Warmer temperature and higher precipitation during summer and winter in Finland are expected to impact ticks in several ways. Higher tick abundance, longer activity seasons, and range expansions of both native and invasive tick species are expected to occur. For example, the invasive tick species Hyalomma marginatum in migratory birds has already been occasionally reported in Finland [9]. In the other northern European countries, the vector of Babesia canis, Dermacentor reticulatus, has been observed in dogs and migratory birds [34].
We encountered difficulties in tick species identification. The duplex PCR that we used [17] was suitable to identify I. ricinus and I. persulcatus but not R. sanguineus. Therefore, we performed ITS2 PCR-based sequencing for all ticks. However, we were able to sequence only 79.5% (272) of the ticks. The reason for failure with the remaining 70 ticks may be suboptimal quality of the extracted DNA. For all sequenced DNAs, no incongruence was found between qPCR and ITS2 PCR-based sequencing. For R. sanguineus, both methods (ITS2 and COX1 PCR-based sequencing) confirmed its identification. Misidentification of tick species is common. The misidentification rate of ticks collected in different countries and assessed by qualified experts has reached 29.6% [35].
Although information on the prevalence of tick-borne infections in companion animals is limited, infections caused by Borrelia, Anaplasma, Babesia, and TBEV have been reported in Northern European countries [5]. In humans, LB and TBE are the most commonly registered tick-borne diseases in the Nordic countries, including Finland. According to national health care registers, the incidence of microbiologically and clinically confirmed human LB cases is increasing [36]. Our results showed a prevalence of 10.5% for B. burgdorferi s.l. in ticks collected from pets in 2020–2021, which is lower than the average prevalence in questing adult ticks of 48.9 ± 8.4% [37]. In Finnish dogs, the seroprevalence of B. burgdorferi is low (2.9%) [12], and no antibodies to B. burgdorferi were detected in cats. Likewise, another seroprevalence study conducted elsewhere in Europe indicated the rarity of B. burgdorferi antibodies in feline samples [38]. Dogs can become infected with B. burgdorferi and develop antibodies, but unlike humans, they rarely get sick. The signs in dogs include fever, fatigue, loss of appetite, intermittent lameness, and swollen and painful joints; skin rash is not observed in animals [39].
In contrast to B. burgdorferi s.l., B. miyamotoi showed a prevalence (1.5%, 95% confidence interval [CI] 0.5–3.4) that is similar to that reported in questing ticks from nationwide studies (0.7%) and in our recent larger collection of ticks from the capital region of Finland (0.6%) [37, 40]. Our previous results also confirmed the circulation of B. miyamotoi in ticks from Finland without the detection of bacterial DNA in a large collection of human samples [40]. However, clinical human infections caused by B. miyamotoi have been reported elsewhere, confirming its association with human, but not pet animal, disease [41].
We found TBEV in only one tick, representing a low prevalence and corresponding with another local study [37]. The most recent nationwide study on a very large collection of ticks did not detect TBEV in ticks from Finland [37], although the previous nationwide study based on crowdsourcing conducted in 2015 showed a prevalence of 0.2% and 3.0% in I. ricinus and I. persulcatus, respectively [37]. Overall, TBEV has a very focal distribution in ticks, and extrapolating over larger areas is uncertain. In dogs, TBEV can cause severe and even fatal neurological symptoms, but the high seroprevalence in healthy dogs in some areas, such as in the Åland Islands, indicates that TBEV results mostly in subclinical infection [13]. Further, dogs can be used as sentinels for TBEV and provide an idea for public health surveillance [42].
We detected A. phagocytophilum DNA in 12 ticks (3.5%, 95% CI 1.8–6.1), of which three infested a single animal. This prevalence is slightly higher than the prevalence reported in questing ticks (0.6%) [7]. The seroprevalence of A. phagocytophilum in Finnish dogs is as high as 5.3%, indicating that infections are common in dogs [12].
Ehrlichia canis can cause a serious disease in dogs and is mainly transmitted by R. sanguineus, which is not endemic in Finland. In this study, the single R. sanguineus tick was imported from Spain, and E. canis was not found. We did not detect E. canis in other ticks either. Its absence corresponds with the low seroprevalence of E. canis (0.3%) in dogs from Finland [12]. Further, the presence of E. canis in the most common tick species from the Nordic countries (Ixodes sp.) has not been confirmed. However, a low prevalence was documented in I. ricinus ticks from the Netherlands [43].
Babesia spp. have medical and veterinary importance. According to the Finnish Food Authority, bovine babesiosis was last reported in 2021 [44]. In Finnish humans, a fatal case due to B. divergens was reported in a previously ill man who was infected simultaneously with Borrelia in 2004 [45]. We are unaware of any other cases at the time of this study. In the current study, B. venatorum was found in ticks (three in I. ricinus and one in R. sanguineus) collected from Taivassalo, Jyväskylä, and Tampere, which are on the southern coast and central part of Finland. Babesia venatorum was detected in Finnish ticks collected in 2015 [30], although no human or animal cases have been reported in the country. However, animal and human infections due to B. venatorum have been reported elsewhere [46, 47].
The vector of Babesia canis, D. reticulatus, has not been reported in Finland. However, canine blood samples have confirmed the presence of B. canis DNA, and reports have confirmed canine babesiosis in imported dogs [48, 49]. Further, other Nordic countries have confirmed the occasional presence of D. reticulatus ticks on dogs, migratory birds, and in nature [34].
In addition to well-known tick-borne pathogens, we studied the prevalence of Ca. N. mikurensis, a bacterium emerging in Europe [50]. This bacterium was recently detected in both I. ricinus and I. persulcatus in Finland [30]. We found a prevalence of 0.9%, which is similar to that reported in I. ricinus (0.8%) [7]. Candidatus N. mikurensis causes disease in immunocompromised humans and has also been detected in a splenectomized dog [50, 51].
Optimally, follow-up samples would have been available from the animals to study transmission and survey the clinical relevance of the microbial findings. However, such samples were not available in this study and may be available in future studies.
The availability of next-generation technologies means that high-throughput sequencing of the full 16S gene is becoming a reality for species and strain-level bacterial detection and could be used by veterinary laboratories for faster detection of a range of pathogens. Despite the wide use of 16S NGS in bacterial detection, the method still has limitations. For example, the quality and quantity of the extracted DNA may affect the results, especially with hard-bodied ticks that have a thick exoskeleton. Further, the presence of different contaminants (from the kits of extraction or the PCR) can confound the generated data [52]. In our study, as also implied by our inability to fully identify the tick species of a proportion of samples, suboptimal quality of DNA may have limited our NGS findings.
In the future, full-length 16S ribosomal RNA gene sequencing of clinical samples may support species identification and provide important information on bacterial community differences between tick species and geographical locations.
Conclusions
The infection rate (17.2%) of ticks in this study and the frequency of infestation with engorged or several ticks highlight the risk of pathogen transmission and the need for specific tests when tick-borne infections are suspected and preparedness to treat potentially more serious clinical diseases if multiple pathogens are transmitted. For proper awareness, tick species, tick abundance, and tick-borne pathogens should be surveyed. Pets can be used as sentinels in this regard. Our results emphasize the usefulness of regular veterinary surveillance and need for continuous monitoring for zoonotic infections in humans and animals. We also recommend screening for both known and novel pathogens when suspecting tick-borne infections in humans and animals to avoid misdiagnosis. To control the entry of novel vector species and pathogens, traveling animals should be treated against ectoparasites.
Availability of data and materials
Sequences of Babesia species (OR167996–OR167999) and tick species identification by ITS2 (I. ricinus OR204991–OR205210; I. persulcatus OR168651–OR168697; R. sanguineus OR168702) and COX1 (R. sanguineus OR167605) genes are deposited in GenBank. Other datasets generated during the current study are not publicly available due to the need to protect the privacy of the animal owners, but they are partially available from the corresponding author on reasonable request.
References
Benelli G. Pathogens Manipulating tick behavior—through a glass, darkly. Pathogens. 2020;9:664. https://doi.org/10.3390/pathogens9080664.
Tahir D, Meyer L, Fourie J, Jongejan F, Mather T, Choumet V, et al. Interrupted blood feeding in ticks: causes and consequences. Microorganisms. 2020;8:910. https://doi.org/10.3390/microorganisms8060910.
Richards SL, Langley R, Apperson CS, Watson E. Do tick attachment times vary between different tick-pathogen systems? Environments. 2017;4:37.
Pfister K, Armstrong R. Systemically and cutaneously distributed ectoparasiticides: a review of the efficacy against ticks and fleas on dogs. Parasit Vectors. 2016;9:436. https://doi.org/10.1186/s13071-016-1719-7.
Cutler SJ, Vayssier-Taussat M, Estrada-Pena A, Potkonjak A, Mihalca AD, Zeller H. Tick-borne diseases and co-infection: current considerations. Ticks Tick Borne Dis. 2021;12:101607. https://doi.org/10.1016/j.ttbdis.2020.101607.
Sanchez-Vicente S, Tagliafierro T, Coleman JL, Benach JL, Tokarz R. Polymicrobial nature of tick-borne diseases. mBio. 2019;10:e02055-e2119. https://doi.org/10.1128/mBio02055-19.
Laaksonen M, Klemola T, Feuth E, Sormunen JJ, Puisto A, Mäkela S, et al. Tick-borne pathogens in Finland: comparison of Ixodes ricinus and I. persulcatus in sympatric and parapatric areas. Parasit Vectors. 2018;11:556. https://doi.org/10.1186/s13071-018-3131-y.
Uusitalo R, Siljander M, Linden A, Sormunen JJ, Aalto J, Hendrickx G, et al. Predicting habitat suitability for Ixodes ricinus and Ixodes persulcatus ticks in Finland. Parasit Vectors. 2022;15:310. https://doi.org/10.1186/s13071-022-05410-8.
Sormunen JJ, Klemola T, Vesterinen EJ. Ticks (Acari: Ixodidae) parasitizing migrating and local breeding birds in Finland. Exp Appl Acarol. 2022;86:145–56. https://doi.org/10.1007/s10493-021-00679-3.
Wright I, Jongejan F, Marcondes M, Peregrine A, Baneth G, Bourdeau P, et al. Parasites and vector-borne diseases disseminated by rehomed dogs. Parasit Vectors. 2020;13:546. https://doi.org/10.1186/s13071-020-04407-5.
Tartuntatautirekisteri [National Infectious Diseases Register]. Finnish Institute for Health and Welfare. 2022. https://www.thl.fi/ttr/gen/rpt/tilastot.html. Accessed 31 Oct 2022.
Pérez Vera C, Kapiainen S, Junnikkala S, Aaltonen K, Spillmann T, Vapalahti O. Survey of selected tick-borne diseases in dogs in Finland. Parasit Vectors. 2014;7:285. https://doi.org/10.1186/1756-3305-7-285.
Levanov L, Pérez Vera C, Vapalahti O. Prevalence estimation of tick-borne encephalitis virus (TBEV) antibodies in dogs from Finland using novel dog anti-TBEV IgG MAb-capture and IgG immunofluorescence assays based on recombinant TBEV subviral particles. Ticks Tick Borne Dis. 2016;7:979–82. https://doi.org/10.1016/j.ttbdis.2016.05.002.
Voyiatzaki C, Papailia SI, Venetikou MS, Pouris J, Tsoumani ME, Papageorgiou EG. Climate changes exacerbate the spread of Ixodes ricinus and the occurrence of Lyme borreliosis and tick-borne encephalitis in Europe—how climate models are used as a risk assessment approach for tick-borne diseases. Int J Environ Res Public Health. 2022;19:6516. https://doi.org/10.3390/ijerph19116516.
Wijnveld M, Schotta A-M, Stelzer T, Duscher G, Leschnik M, Stockinger H, et al. Novel protozoans in Austria revealed through the use of dogs as sentinels for ticks and tick-borne pathogens. Microorganisms. 2021;9:1392. https://doi.org/10.3390/microorganisms9071392.
Protect Our Future Too project. MSD Animal Health. 2023. www.protectourfuturetoo.com. Accessed 6 Mar 2023.
Sormunen JJ, Penttinen R, Klemola T, Hänninen J, Vuorinen I, Laaksonen M, et al. Tick-borne bacterial pathogens in southwestern Finland. Parasit Vectors. 2016;9:168. https://doi.org/10.1186/s13071-016-1449-x.
Rumer L, Sheshukova O, Dautel H, Donoso Mantke O, Niedrig M. Differentiation of medically important Euro-Asian tick species Ixodes ricinus, Ixodes persulcatus, Ixodes hexagonus, and Dermacentor reticulatus by polymerase chain reaction. Vector Borne Zoonotic Dis. 2011;11:899–905. https://doi.org/10.1089/vbz.2009.0191.
Kushimo OM. The tick genus Amblyomma in Africa: phylogeny and mutilocus DNA barcoding. [Electronic Thesis and Dissertation]. Statesboro, United States Georgia Southern University; 2013.
Zakham F, Albalawi AE, Alanazi AD, Truong Nguyen P, Alouffi AS, Alaoui A, et al. Viral RNA metagenomics of Hyalomma ticks collected from dromedary camels in Makkah province, Saudi Arabia. Viruses. 2021;13:1396. https://doi.org/10.3390/v13071396.
Schwaiger M, Cassinotti P. Development of a quantitative real-time RT-PCR assay with internal control for the laboratory detection of tick borne encephalitis virus (TBEV) RNA. J Clin Virol. 2003;27:136–45.
Song J, Zhao S, Li Y, Wang H, Zhang L, Wang J, et al. Duplex TaqMan real-time PCR assay for simultaneous detection and quantification of Anaplasma capra and Anaplasma phagocytophilum infection. Mol Cell Probes. 2020;49:101487. https://doi.org/10.1016/j.mcp.2019.101487.
Georges K, Loria GR, Riili S, Greco A, Caracappa S, Jongejan F, et al. Detection of haemoparasites in cattle by reverse line blot hybridisation with a note on the distribution of ticks in Sicily. Vet Parasitol. 2001;99:273–86. https://doi.org/10.1016/s0304-4017(01)00488-5.
Keegan KP, Glass EM, Meyer F. MG-RAST, a metagenomics service for analysis of microbial community structure and function. Methods Mol Biol. 2016;1399:207–33. https://doi.org/10.1007/978-1-4939-3369-3_13.
Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. https://doi.org/10.1128/AEM.00062-07.
Klitgaard K, Kjaer LJ, Isbrand A, Hansen MF, Bødker R. Multiple infections in questing nymphs and adult female Ixodes ricinus ticks collected in a recreational forest in Denmark. Ticks Tick Borne Dis. 2019;10:1060–5.
del Cerro A, Oleaga A, Somoano A, Barandika JF, Garcia-Perez AL, Espi A. Molecular identification of tick-borne pathogens (Rickettsia spp., Anaplasma phagocytophilum, Borrelia burgdorferi sensu lato, Coxiella burnetii and piroplasms) in questing and feeding hard ticks from North-Western Spain. Ticks Tick Borne Dis. 2022;13:101961.
Nyrhilä S, Sormunen JJ, Mäkelä S, Sippola E, Vesterinen EJ, Klemola T. One out of ten: low sampling efficiency of cloth dragging challenges abundance estimates of questing ticks. Exp Appl Acarol. 2020;82:571–85. https://doi.org/10.1007/s10493-020-00564-5.
Overzier E, Pfister K, Herb I, Mahling M, Bock G, Silaghi C. Detection of tick-borne pathogens in roe deer (Capreolus capreolus), in questing ticks (Ixodes ricinus), and in ticks infesting roe deer in southern Germany. Ticks Tick Borne Dis. 2013;4:320–8.
Laaksonen M, Sajanti E, Sormunen JJ, Penttinen R, Hänninen J, Ruohomäki K, et al. Crowdsourcing-based nationwide tick collection reveals the distribution of Ixodes ricinus and I. persulcatus and associated pathogens in Finland. Emerg Microbes Infect. 2017;6:e31. https://doi.org/10.1038/emi.2017.17.
Kovalev SY, Golovljova IV, Mukhacheva TA. Natural hybridization between Ixodes ricinus and Ixodes persulcatus ticks evidenced by molecular genetics methods. Ticks Tick Borne Dis. 2016;7:113–8.
Schafer I, Volkmann M, Beelitz P, Merle R, Müller E, Kohn B. Retrospective analysis of vector-borne infections in dogs after travelling to endemic areas (2007–2018). Vet Parasitol. 2019;276:100015.
Ruosteenoja K, Jylhä K. Projected climate change in Finland during the 21st century calculated from CMIP6 model simulations. Geophysica. 2022;56:39–69.
Kjaer LJ, Soleng A, Edgar KS, Lindstedt HEH, Paulsen KM, Andreassen ÅK, et al. A large-scale screening for the taiga tick, Ixodes persulcatus, and the meadow tick, Dermacentor reticulatus, in southern Scandinavia, 2016. Parasit Vectors. 2019;12:338. https://doi.org/10.1186/s13071-019-3596-3.
Estrada-Peña A, D’Amico G, Palomar AM, Dupraz M, Fonville M, Heylen D, et al. A comparative test of ixodid tick identification by a network of European researchers. Ticks Tick Borne Dis. 2017;8:540–6. https://doi.org/10.1016/j.ttbdis.2017.03001.
Feuth E, Virtanen M, Helve O, Hytönen J, Sane J. Lyme borreliosis in Finland: a register-based linkage study. BMC Infect Dis. 2020;20:819.
Sormunen JJ, Andersson T, Aspi J, Back J, Cederberg T, Haavisto N, et al. Monitoring of ticks and tick-borne pathogens through a nationwide research station network in Finland. Ticks Tick Borne Dis. 2020;11:101449. https://doi.org/10.1016/j.ttbdis.2020.101449.
Pantchev N, Vrhovec MG, Pluta S, Straubinger RK. Seropositivity of Borrelia burgdorferi in a cohort of symptomatic cats from Europe based on a C6-peptide assay with discussion of implications in disease aetiology. Berl Munch Tierarztl Wochenschr. 2016;129:333–9.
Vogt NA. Lyme borreliosis in animals. In: Merck Veterinary Manual. 2022. www.merckvetmanual.com/generalized-conditions/lyme-borreliosis/lyme-borreliosis-in-animals. Accessed 31 Oct 2022.
Zakham F, Jääskeläinen AJ, Castrén J, Sormunen JJ, Uusitalo R, Smura T, et al. Molecular detection and phylogenetic analysis of Borrelia miyamotoi strains from ticks collected in the capital region of Finland. Ticks Tick Borne Dis. 2021;12:101608. https://doi.org/10.1016/j.ttbdis.2020.101608.
Hoornstra D, Azagi T, van Eck JA, Wagemakers A, Koetsveld J, Spijker R, et al. Prevalence and clinical manifestation of Borrelia miyamotoi in Ixodes ticks and humans in the northern hemisphere: a systematic review and meta-analysis. Lancet Microbe. 2022;3:e772–86. https://doi.org/10.1016/S2666-5247(22)00157-4.
Pfeffer M, Dobler G. Tick-borne encephalitis virus in dogs—is this an issue? Parasit Vectors. 2011;4:59. https://doi.org/10.1186/1756-3305-4-59.
Wielinga PR, Gaasenbeek C, Fonville M, de Boer A, de Vries A, Dimmers W, et al. Longitudinal analysis of tick densities and Borrelia, Anaplasma, and Ehrlichia infections of Ixodes ricinus ticks in different habitat areas in The Netherlands. Appl Environ Microbiol. 2006;72:7594–601. https://doi.org/10.1128/AEM.01851-06.
Finnish Food Authority. Eläintaudit Suomessa 2021 [Animal diseases in Finland, 2021]. Ruokaviraston julkaisuja 2022:4. http://hdl.handle.net/10138/347520. Accessed 12 Dec 2022.
Haapasalo K, Suomalainen P, Sukura A, Siikamaki H, Jokiranta TS. Fatal babesiosis in man, Finland, 2004. Emerg Infect Dis. 2010;16:1116–8. https://doi.org/10.3201/eid1607.091905.
Gray A, Capewell P, Loney C, Katzer F, Shiels BR, Weir W. Sheep as host species for zoonotic Babesia venatorum. United Kingdom Emerg Infect Dis. 2019;25:2257–60. https://doi.org/10.3201/eid2512.190459.
Sun Y, Li SG, Jiang JF, Wang X, Zhang Y, Wang H, et al. Babesia venatorum infection in child. China Emerg Infect Dis. 2014;20:896–7. https://doi.org/10.3201/eid2005.121034.
Bajer A, Beck A, Beck R, Behnke JM, Dwuznik-Szarek D, Eichenberger RM, et al. Babesiosis in Southeastern, Central and Northeastern Europe: an emerging and re-emerging tick-borne disease of humans and animals. Microorganisms. 2022;10:945. https://doi.org/10.3390/microorganisms10050945.
Birkenheuer AJ, Buch J, Beall MJ, Braff J, Chandrashekar R. Global distribution of canine Babesia species identified by a commercial diagnostic laboratory. Vet Parasitol. 2020;22:100471.
Portillo A, Santibanez P, Palomar AM, Santibanez S, Oteo JA. Candidatus Neoehrlichia mikurensis in Europe. New Microbes New Infect. 2018;22:30–6.
Hofmann-Lehmann R, Wagmann N, Meli ML, Riond B, Novacco M, Joekel D, et al. Detection of 'Candidatus Neoehrlichia mikurensis’ and other Anaplasmataceae and Rickettsiaceae in Canidae in Switzerland and Mediterranean countries. Schweiz Arch Tierheilkd. 2016;158:691–700. https://doi.org/10.17236/sat00087.
Couper L, Swei A. Tick microbiome characterization by next-generation 16S rRNA amplicon sequencing. J Vis Exp. 2018;138:58239. https://doi.org/10.3791/58239.
Ivacic L, Reed KD, Mitchell PD, Ghebranious N. A LightCycler TaqMan assay for detection of Borrelia burgdorferi sensu lato in clinical samples. Diagn Microbiol Infect Dis. 2007;57:137–43. https://doi.org/10.1016/jdiagmicrobio.2006.08.005.
Hovius JW, de Wever B, Sohne M, Brouwer MC, Coumou J, Wagemakers A, et al. A case of meningoencephalitis by the relapsing fever spirochaete Borrelia miyamotoi in Europe. Lancet. 2013;382:658. https://doi.org/10.1016/S0140-6736(13)61644-X.
Jahfari S, Coipan EC, Fonville M, van Leeuwen AD, Hengeveld P, Heylen D, et al. Circulation of four Anaplasma phagocytophilum ecotypes in Europe. Parasit Vectors. 2014;7:365. https://doi.org/10.1186/1756-3305-7-365.
Øines Ø, Radzijevskaja J, Paulauskas A, Rosef O. Prevalence and diversity of Babesia spp. in questing Ixodes ricinus ticks from Norway. Parasit Vectors. 2012;5:156. https://doi.org/10.1186/1756-3305-5-156.
Jahfari S, Fonville M, Hengeveld P, Reusken C, Scholte EJ, Takken W, et al. Prevalence of Neoehrlichia mikurensis in ticks and rodents from North-West Europe. Parasit Vectors. 2012;5:74. https://doi.org/10.1186/1756-3305-5-74.
Hornok S, Muhldorfer K, Takacs N, Hofmann-Lehmann R, Meli ML, Gyuranecz M, et al. Broad range screening of vector-borne pathogens in arctic foxes (Vulpes lagopus) in Iceland. Animals (Basel). 2020;10:2031. https://doi.org/10.3390/ani10112031.
Acknowledgements
The authors would like to thank Johanna Martikainen and Eetu Sironen for their help in reporting the data and MSD partners Melina Bruno-Paasisalo, Risto Pulkkinen, Eppu Petrelius, Minna Lahti, and Randi Lintrup for supporting this study. We also thank our partners Dr Jani Sormunen from the University of Turku, Finland, and Tomas Jinnerot at National Veterinary Institute, Sweden, for providing positive controls. We thank all the participating clinics and pet owners.
Funding
Open Access funding provided by University of Helsinki including Helsinki University Central Hospital. This study was funded by MSD Animal Health and supported by the Veclimit Academy of Finland (decision #329323), CrossBar Academy of Finland (decision #339510), and Jane and Aatos Erkko Foundation. Except for PMK as a co-author (see Competing interests and Authors’ contributions), the actual funding bodies have not participated in the design of the study, the collection, analysis, or interpretation of the data or writing the manuscript.
Author information
Authors and Affiliations
Contributions
FZ, PTP, and RSC performed experimental work. FZ analysed the results and wrote the first draft of the manuscript. EMK and TSi planned and coordinated this study. RU designed the map and analysed geographical data. TSm performed NGS. RK performed NGS analysis of results. PMK, OV, and TSi planned the general approach of the study. PMK organized sample collection. All authors reviewed drafts and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The owners of the animals and the veterinary clinics gave their informed consent to voluntarily participate in this study.
Consent for publication
Not applicable.
Competing interests
PMK is an employee of MSD Animal Health in addition to having the title of Adjunct Professor in the University of Helsinki. The other authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Comparison between qPCR and 16S next-generation sequencing results.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zakham, F., Korhonen, E.M., Puonti, P.T. et al. Molecular detection of pathogens from ticks collected from dogs and cats at veterinary clinics in Finland. Parasites Vectors 16, 327 (2023). https://doi.org/10.1186/s13071-023-05864-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-023-05864-4