Abstract
Background
Several species belonging to the genus Ehrlichia are considered pathogenic for animals and humans. Although wildlife are known to play an important role in the epidemiology of these bacteria, information on the role of wild lagomorphs in their sylvatic cycle is limited. Thus, the objective of the present study was to assess the occurrence of Ehrlichia spp. in ticks collected from wild lagomorphs in Spanish Mediterranean ecosystems.
Methods
A total of 1122 pooled ticks (254 pools) collected from 506 wild rabbits (Oryctolagus cuniculus) and 29 Iberian hares (Lepus granatensis) were analysed using a nested PCR assay targeting the partial groEL gene. Ehrlichia spp.-positive samples were further subjected to a second PCR assay targeting 16S rRNA.
Results
Three (1.2%) tick pools comprising Rhipicephalus pusillus collected from nine wild rabbits were positive for Ehrlichia spp. All the Ehrlichia DNA sequences were identical, and use of sequence and phylogenetic analyses allowed us to identify a novel Ehrlichia species.
Conclusions
We provide evidence that a novel Ehrlichia species, named herein as ‘Candidatus Ehrlichia andalusi’, which may be of concern for animal and public health, is circulating in R. pusillus in Spanish Mediterranean ecosystems. Further studies are warranted to assess the epidemiology, pathogenicity and zoonotic potential of this Ehrlichia species.
Graphical Abstract

Similar content being viewed by others
Background
The incidence of tick-borne pathogens has increased worldwide during the last decades [1]. This emergence, or re-emergence, may be related to climate change, global travel, changes in land use (urbanization, deforestation, habitat fragmentation, etc.), or an increase in outdoor activities, among other factors [2]. Scientists and health authorities are very concerned about tick-borne agents of disease, so increased diagnostic pressure may also explain the increased detection of these pathogens in vectors, other animals and humans [3].
Among the wide variety of tick-borne pathogens, those belonging to the family Anaplasmataceae are of special interest due to their zoonotic potential and worldwide distribution [4]. Within this family, the genus Ehrlichia is of major concern. Several species belonging to this genus are considered pathogenic for both domestic and wild animals, such as Ehrlichia canis, Ehrlichia chaffeensis and Ehrlichia ruminantium [5]. In addition, Ehrlichia canis, Ehrlichia chaffeensis, Ehrlichia ewingii and Ehrlichia muris have been shown to be zoonotic [6].
Since transovarial transmission of Ehrlichia spp. has not been demonstrated in ticks [7], it has been suggested that wildlife may play an important role in the epidemiology of these pathogens [8]. Although there is an increasing number of studies providing information on the presence and prevalence of Ehrlichia spp. in domestic and wild ruminants and their ticks, data on the epidemiology of these bacteria in ticks collected from wild lagomorphs are still scarce. Therefore, the aim of the present study was to molecularly determine the occurrence of Ehrlichia spp. in pools of ticks parasitizing wild rabbits (Oryctolagus cuniculus) and Iberian hares (Lepus granatensis) in Mediterranean ecosystems in southern Spain.
Methods
Sample collection
Between October 2016 and August 2020, a total of 1122 ticks were collected from 506 wild rabbits (total number of rabbits examined = 1304) and 29 Iberian hares (total number of hares examined = 58). These specimens were identified in a previous study [9] as Rhipicephalus pusillus, Rhipicephalus sanguineus sensu lato, Haemaphysalis hispanica, Hyalomma lusitanicum and Ixodes ventalloi. The ticks were kept frozen at − 20 °C until examination.
For the detection of Ehrlichia spp. DNA, ticks collected from wild rabbits and Iberian hares hunted in the same hunting area were pooled according to species, development stage and host species [9]. The number of pools for each tick species is summarized in Table 1.
Molecular analyses
Tick DNA was extracted using a commercial kit (High Pure PCR Template Preparation Kit; Roche Diagnostics, Mannheim, Germany), following the manufacturer’s instructions. Ehrlichia spp. DNA was detected by a nested PCR assay targeting a partial fragment of the groEL gene [10, 11] Amplicons of the expected size were purified, sequenced, aligned and edited as previously reported [9]; consensus sequences were then scanned against the GenBank database using the Basic Local Alignment Search Tool. All Ehrlichia spp.-positive samples were further subjected to a second PCR protocol targeting the 16S rRNA of these bacteria [5, 12, 13]. The PCR products were processed, sequenced and analysed again, as previously described.
A phylogenetic analysis was carried out using MrBayes 3.2.7 software [14] by Bayesian approach with Markov Chain Monte Carlo sampling (10,000,000 generations sampling every 1000 steps). A Hasegawa-Kishino-Yano substitution model with gamma-distributed rate variation across sites was used for the analysis of Ehrlichia spp. sequences at the groEL and 16s rRNA genes. The model was selected based on Akaike information criterion values using the free software jModelTest v.2.1.10 [15]. The tree was visualized and edited using FigTree 1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Statistical analysis
Maximum likelihood estimation was used to estimate the prevalence of Ehrlichia spp. in pooled R. pusillus [16]. Statistical analyses were performed using the statistical software R 4.2.1 [17] and the functions llprevr and dprev [16].
Results and discussion
Only three out of the 254 (1.2%) tick pools (maximum likelihood estimate 0.3%, and 95% confidence interval 0.1–0.7) yielded positive results with respect to targeting of the groEL partial gene (Table 1). These results revealed that Ehrlichia spp. were not prevalent in the ticks collected from the wild lagomorphs from Mediterranean ecosystems of southern Spain, which suggests that these ticks probably do not play an important role in the sylvatic cycle of these pathogens.
All the positive pools comprised female R. pusillus obtained from nine rabbits hunted in three hunting areas in eastern and western Andalusia (Fig. 1), and represent, to the best of our knowledge, the first report of Ehrlichia spp. in R. pusillus. To the best of our knowledge, there is only one previous report of Anaplasmataceae in this tick species, where 1.8% of R. pusillus collected from horses in France were found to be positive for Anaplasma phagocytophilum [18]. Ehrlichia DNA was not detected in the other tick species collected from the wild rabbits or the Iberian hares. However, R. sanguineus sensu lato is known to be involved in the transmission of numerous pathogens, including E. canis [19], and questing I. ventalloi from Portugal and Spain were found to harbour Anaplasmataceae, including A. marginale and A. phagocytophilum [20,21,22]. The vectorial competence of H. lusitanicum in the transmission of Ehrlichia spp. is poorly understood; however, DNA of these bacteria was detected in H. lusitanicum from Italy [23]. Finally, little is known about tick-borne pathogens in H. hispanica [24].
The sequences identified in this study were deposited in GenBank under accession numbers OP490270 and OP502086. Sequence analysis revealed that all the Ehrlichia spp. isolates were identical to each other at both the groEL and 16S rRNA genes. For the groEL gene, all the sequences had a percentage nucleotide identity between 91.5% to 91.7% when compared to uncultured Ehrlichia sp. clone Tajikistan sequences KJ930191 and KJ930192 obtained from Hyalomma anatolicum from Tajikistan [25]; 91.7% identity with sequences MW054555 and MW054557 deposited for Ehrlichia sp. isolate YNT obtained from Rhipicephalus annulatus and Rhipicephalus geigyi from Guinea [26] was also found. In addition, the nucleotide sequences at the 16S rRNA partial gene showed a percentage identity ranging from 99.4 to 99.7% when compared to several deposited sequences of uncultured Ehrlichia spp. (AF311968, AY309970, KJ410257, KX987325, KX577724, KY046298, MH250197, MT258392 and OK481113) from different species of Hyalomma, Rhipicephalus and Haemaphysalis from African and Asian countries, including Angola [27], China [28, 29], Japan [30, 31], Malaysia [32], Niger [33] and Pakistan [34]. The degree of similarity between the Ehrlichia species at the 16S rRNA gene could indicate that it is not an appropriate gene for discriminating between species of this genus, similar to previous conclusions for other bacterial genera [35].
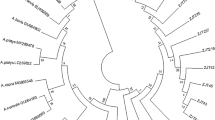
Phylogenetic trees constructed with partial sequences of the groEL and 16S rRNA genes had similar topologies (Figs. 2, 3). The groEL sequences formed a clade with sequence KJ930194 detected in H. anatolicum from Tajikistan [25], which was clearly separate from the main Ehrlichia species. Similarly, the 16S rRNA sequences of Ehrlichia sp. obtained in this study formed a clade with sequence JX402605 obtained from Hyalomma asiaticum from China [28]. The genetic distances and phylogenetic relationships indicated that a novel Ehrlichia species had been found, which is named herein as ‘Candidatus Ehrlichia andalusi’. Interestingly, the positive pools comprised ticks collected from wild rabbits from different hunting areas in eastern and western Andalusia (Fig. 1). Since wild rabbits are territorial and live close to their warrens, and their home range is not larger than 10 ha [36], the detection of this novel species in ticks from three geographically separated wild rabbit populations suggests that it may be distributed throughout southern Spain. In support of this hypothesis, no translocations of wild rabbits have been carried out in these hunting areas according to data collected by the gamekeepers.
Phylogenetic tree clustering of the partial groEL of Ehrlichia spp. The tree was obtained using a Hasegawa–Kishino–Yano substitution model with gamma-distributed rate variation across sites (HKY + G) with the software MrBayes 3.2.7 [14] by Bayesian approach with Markov Chain Monte Carlo sampling (10,000,000 generations sampling every 1000 steps). This analysis involved 47 nucleotide sequences. The nucleotide sequence of Neoehrlichia mikurensis was used as an outgroup. The isolate identified in this study is indicated in bold
Phylogenetic tree clustering of the partial 16S RNA of Ehrlichia spp. The tree was obtained using HKY + G with the software MrBayes 3.2.7 [14] by Bayesian approach with Markov Chain Monte Carlo sampling (10,000,000 generations sampling every 1000 steps). This analysis involved 66 nucleotide sequences. The nucleotide sequence of Neoehrlichia mikurensis was used as an outgroup. The isolate identified in this study is indicated in bold
Novel Ehrlichia species and strains have been reported worldwide during the last decades, suggesting that there are several knowledge gaps in the epidemiology and phylogeny of these zoonotic bacteria, especially regarding their sylvatic cycles. Most of these novel organisms were reported for ticks that feed on both domestic and wild animals in South American [37,38,39,40,41,42,43] and Asian countries [28, 30, 32, 44]. However, reports of novel Ehrlichia species are very scare for Europe, and mainly restricted to ticks collected from wild animals. Ehrlichia sp. HF strain was detected in Ixodes ricinus collected from the European wood mouse (Apodemus sylvaticus) in France [45], as well as in Ixodes apronophorus, Ixodes ricinus and R. sanguineus collected from dogs and foxes in Romania [46, 47]. In addition, a strain similar to Ehrlichia chaffeensis and Ehrlichia muris was detected in song thrush (Turdus philomelos) from Hungary [48].
Since all developmental stages of R. pusillus are known to feed on lagomorphs, especially wild rabbits [49, 50], this tick may have a restricted host range. However, it has been sporadically reported in other mammals, such as rodents, ungulates, carnivores and humans [49]. Considering that no transovarial transmission of Ehrlichia spp. has been reported in ticks [7], the detection of ‘Candidatus E. andalusi’ in R. pusillus that were feeding on rabbits may be an accidental finding that is not related to lagomorph populations. Unfortunately, as we were unable to obtain tissue or blood samples from the hunted wild rabbits and hares, we were unable to further examine the role of these lagomorph species in the epidemiology of this pathogen. In this regard, future studies are warranted to investigate the presence of this pathogen in populations of hosts of R. pusillus.
Although several Ehrlichia species are considered to be pathogenic for humans and animals [6, 51], information on their presence in host or vector populations in Europe is limited. Our results suggest that Ehrlichia species of unknown pathogenicity are circulating in wild animal populations or in the ticks that they harbour, which may be of concern for human and animal health. Further studies are needed to determine the presence, prevalence and reservoir range of the Ehrlichia species present in Mediterranean ecosystems, and to unravel their epidemiology, pathogenicity and phylogenetic relationships.
Availability of data and materials
The data that support the findings of this study are available from the authors upon reasonable request.
References
Diuk-Wasser MA, VanAcker MC, Fernández MP. Impact of land use changes and habitat fragmentation on the eco-epidemiology of tick-borne diseases. J Med Entomol. 2021;58:1546–64. https://doi.org/10.1093/jme/tjaa209.
Ortíz DI, Piche-Ovares M, Romero-Vega LM, Wagman J, Troyo A. The impact of deforestation, urbanization, and changing land use patterns on the ecology of mosquito and tick-borne diseases in Central America. Insects. 2022;13:20. https://doi.org/10.3390/insects13010020.
Inci A, Yildirim A, Duzlu O, Doganay M, Aksoy S. Tick-borne diseases in Turkey: a review based on One Health perspective. PLOS Negl Trop Dis. 2016;10:e0005021. https://doi.org/10.1371/journal.pntd.0005021.
André MR. Diversity of Anaplasma and Ehrlichia/Neoehrlichia agents in terrestrial wild carnivores worldwide: implications for human and domestic animal health and wildlife conservation. Front Vet Sci. 2018;5:627654. https://doi.org/10.3389/fvets.2018.00293.
Parola P, Roux V, Camicas JL, Baradji I, Brouqui P, Raoult D. Detection of Ehrlichiae in African ticks by polymerase chain reaction. Trans R Soc Trop Med Hyg. 2000;94:707–8. https://doi.org/10.1016/s0035-9203(00)90243-8.
Lin M, Xiong Q, Chung M, Daugherty SC, Nagaraj S, Sengamalay N, et al. Comparative analysis of genome of Ehrlichia sp. HF, a model bacterium to study fatal human ehrlichiosis. BMC Genom. 2021;22:11. https://doi.org/10.1186/s12864-020-07309-z.
Ismail N, McBride JW. Tick-borne emerging infections ehrlichiosis and anaplasmosis. Clin Lab Med. 2017;37:317–40. https://doi.org/10.1016/j.cll.2017.01.006.
García-Pérez AL, Oporto B, Espí A, del Cerro A, Barral M, Povedano I, et al. Anaplasmataceae in wild ungulates and carnivores in northern Spain. Ticks Tick Borne Dis. 2016;7:264–9. https://doi.org/10.1016/j.ttbdis.2015.10.019.
Remesar S, Castro-Scholten S, Cano-Terriza D, Diaz P, Morrondo P, Jimenez-Martin D, et al. Molecular identification of zoonotic Rickettsia species in Ixodidae parasitizing wild lagomorphs from Mediterranean ecosystems. Transbound Emerg Dis. 2021;69:e992–1004. https://doi.org/10.1111/tbed.14379.
Nicholson WL, Castro MB, Kramer VL, Sumner JW, Childs JE. Dusky-footed wood rats (Neotoma fuscipes) as reservoirs of granulocytic Ehrlichiae (Rickettsiales: Ehrlichieae) in northern California. J Clin Microbiol. 1999;37:3323–7. https://doi.org/10.1128/jcm.37.10.3323-3327.1999.
Mongruel AC, Benevenute JL, André MR, Carrasco AO, Machado RZ, Seki MC. Molecular characterization of Anaplasma sp. in free-living gray brockets (Mazama gouazoubira). Vector Borne Zoonotic Dis. 2017;17:165–71. https://doi.org/10.1089/vbz.2016.2026.
Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. https://doi.org/10.1128/jb.173.2.697-703.1991.
Inokuma H, Beppu T, Okuda M, Shimada Y, Sakata Y. Detection of ehrlichial DNA in Haemaphysalis ticks recovered from dogs in Japan that is closely related to a novel Ehrlichia sp. found in cattle ticks from Tibet, Thailand, and Africa. J Clin Microbiol. 2004;42:1353–5. https://doi.org/10.1128/jcm.42.3.1353-1355.2004.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42. https://doi.org/10.1093/sysbio/sys029.
Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. https://doi.org/10.1038/nmeth.2109.
Williams CJ, Moffitt CM. Estimation of pathogen prevalence in pooled samples using maximum likelihood methods and open-source software. J Aquat Anim Health. 2005;17:386–91. https://doi.org/10.1577/h04-066.1.
R Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria; 2022. https://www.R-project.org/.
Dugat T, Leblond A, Keck N, Lagree AC, Desjardins I, Joulie A, et al. One particular Anaplasma phagocytophilum ecotype infects cattle in the Camargue. France Parasit Vectors. 2017;10:371. https://doi.org/10.1186/s13071-017-2305-3.
Dantas-Torres F. The brown dog tick, Rhipicephalus sanguineus (Latreille, 1806) (Acari:Ixodidae): from taxonomy to control. Vet Parasitol. 2008;152:173–85. https://doi.org/10.1016/j.vetpar.2007.12.030.
Márquez FJ. Spotted fever group Rickettsia in ticks from southeastern Spain natural parks. Exp Appl Acarol. 2008;45:185–94. https://doi.org/10.1007/s10493-008-9181-7.
Antunes S, Ferrolho J, Domingues N, Santos AS, Santos-Silva MM, Domingos A. Anaplasma marginale and Theileria annulata in questing ticks from Portugal. Exp Appl Acarol. 2016;70:79–88. https://doi.org/10.1007/s10493-016-0057-y.
Santos AS, de Bruin A, Veloso AR, Marques C, da Fonseca IP, de Sousa R, et al. Detection of Anaplasma phagocytophilum, Candidatus Neoehrlichia sp., Coxiella burnetii and Rickettsia spp. in questing ticks from a recreational park, Portugal. Ticks Tick Borne Dis. 2018;9:1555–64. https://doi.org/10.1016/j.ttbdis.2018.07.010.
Torina A, Galindo RC, Vicente J, Di Marco V, Russo M, Aronica V, et al. Characterization of Anaplasma phagocytophilum and A. ovis infection in a naturally infected sheep flock with poor health condition. Trop Anim Health Prod. 2010;42:1327–31. https://doi.org/10.1007/s11250-010-9580-8.
Estrada-Peña A, Mihalca AD, Petney T. Ticks of Europe and North Africa. A guide to species identification: Springer International Publishing; 2017.
Kartashov MY, Kononova YV, Petrova ID, Tupota NL, Mikryukova TP, Ternovoi VA, et al. Detection of Ehrlichia spp. and Theileria spp. in Hyalomma anatolicum ticks collected in Tajikistan. Vavilovskii Zh Genet Sel. 2020;24:55–9. https://doi.org/10.18699/vj20.595.
Makenov MT, Toure AH, Korneev MG, Sacko N, Porshakov AM, Yakovlev SA, et al. Rhipicephalus microplus and its vector-borne haemoparasites in Guinea: further species expansion in West Africa. Parasitol Res. 2021;120:1563–70. https://doi.org/10.1007/s00436-021-07122-x.
Palomar AM, Molina I, Bocanegra C, Portillo A, Salvador F, Moreno M, et al. Old zoonotic agents and novel variants of tick-borne microorganisms from Benguela (Angola), July 2017. Parasit Vectors. 2022;15:140. https://doi.org/10.1186/s13071-022-05238-2.
Kang Y-J, Diao X-N, Zhao G-Y, Chen M-H, Xiong Y, Shi M, et al. Extensive diversity of Rickettsiales bacteria in two species of ticks from China and the evolution of the Rickettsiales. BMC Evol Biol. 2014;14:167. https://doi.org/10.1186/s12862-014-0167-2.
Lu M, Tian J-H, Yu B, Guo W-P, Holmes EC, Zhang Y-Z. Extensive diversity of Rickettsiales bacteria in ticks from Wuhan. China Ticks Tick Borne Dis. 2017;8:574–80. https://doi.org/10.1016/j.ttbdis.2017.03.006.
Inokuma H, Brouqui P, Drancourt M, Raoult D. Citrate synthase gene sequence: a new tool for phylogenetic analysis and identification of Ehrlichia. J Clin Microbiol. 2001;39:3031–9. https://doi.org/10.1128/jcm.39.9.3031-3039.2001.
Su H, Onoda E, Tai H, Fujita H, Sakabe S, Azuma K, et al. Diversity unearthed by the estimated molecular phylogeny and ecologically quantitative characteristics of uncultured Ehrlichia bacteria in Haemaphysalis ticks. Japan Sci Rep. 2021;1:687. https://doi.org/10.1038/s41598-020-80690-7.
Koh FX, Kho KL, Kisomi MG, Wong LP, Bulgiba A, Tan PE, et al. Ehrlichia and Anaplasma infections: serological evidence and tick surveillance in Peninsular Malaysia. J Med Entomol. 2018;55:269–76. https://doi.org/10.1093/jme/tjx204.
Parola P, Inokuma H, Camicas JL, Brouqui P, Raoult D. Detection and identification of spotted fever group Rickettsiae and Ehrlichiae in African ticks. Emerg Infect Dis. 2001;7:1014–7. https://doi.org/10.3201/eid0706.010616.
Rehman A, Conraths FJ, Sauter-Louis C, Kruecken J, Nijhof AM. Epidemiology of tick-borne pathogens in the semi-arid and the arid agro-ecological zones of Punjab province. Pakistan Transbound Emerg Dis. 2019;66:526–36. https://doi.org/10.1111/tbed.13059.
Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45:2761–4. https://doi.org/10.1128/JCM.01228-07.
Delibes-Mateos M, Rodel HG, Rouco C, Alves C, Carneiro M, Villafuerte R. European rabbit Oryctolagus cuniculus Linnaeus, 1758. In: Hackl ̈ander, Klaus, Alves, Paulo Celio (Eds.), Handbook of the mammals of Europe: primates and Lagomorpha. Springer Nature; 2021. p. 220 (ISBN 978–3–030–34042–1).
Cabezas-Cruz A, Zweygarth E, Vancova M, Broniszewska M, Grubhoffer L, Friche Passos LM, et al. Ehrlichia minasensis sp nov., isolated from the tick Rhipicephalus microplus. Int J Syst Evol Microbiol. 2016;66:1426–30; doi: https://doi.org/10.1099/ijsem.0.000895.
Cicuttin GL, De Salvo MN, Nava S. Two novel Ehrlichia strains detected in Amblyomma tigrinum ticks associated to dogs in peri-urban areas of Argentina. Comp Immunol Microbiol Infect Dis. 2017;53:40–4. https://doi.org/10.1016/j.cimid.2017.07.001.
Monje LD, Fernandez C, Percara A. Detection of Ehrlichia sp. strain San Luis and Candidatus Rickettsia andeanae in Amblyomma parvum ticks. Ticks Tick-Borne Dis. 2019;10:111–4. https://doi.org/10.1016/j.ttbdis.2018.09.008.
Muñóz-Leal S, Clemes YS, Lopes MG, Acosta ICL, Serpa MCA, Mayorga LFSP, et al. Novel Ehrlichia sp. detected in Magellanic penguins (Sphenicus magellanicus) and in the seabird tick Ixodes uriae from Magdalena Island, southern Chile. Ticks Tick Borne Dis. 2019;10:101256. https://doi.org/10.1016/j.ttbdis.2019.06.015.
Cicuttin GL, De Salvo MN, Perez PD, Silva D, Felix ML, Venzal JM, et al. A novel Ehrlichia strain (Rickettsiales: Anaplasmataceae) detected in Amblyomma triste (Acari: Ixodidae), a tick species of public health importance in the Southern Cone of America. Pathog Glob Health. 2020;114:318–22. https://doi.org/10.1080/20477724.2020.1795579.
Eberhardt AT, Fernandez C, Fargnoli L, Beldomenico PM, Monje LD. A putative novel strain of Ehrlichia infecting Amblyomma tigrinum associated with Pampas fox (Lycalopex gymnocercus) in Esteros del Ibera ecoregion, Argentina. Ticks Tick Borne Dis. 2020;11:101318. https://doi.org/10.1016/j.ttbdis.2019.101318.
Félix LM, Muñóz-Leal S, Andrés Carvalho L, Queirolo D, Remesar S, Teresa Armúa-Fernández M, et al. Characterization of “Candidatus Ehrlichia pampeana”in Haemaphysalis juxtakochi ticks and gray brocket deer (Mazama gouazoubira) from Uruguay. Microorganisms. 2021;9:2165. https://doi.org/10.3390/microorganisms9102165.
Wen BH, Jian R, Zhang YZ, Chen R. Simultaneous detection of Anaplasma marginale and a new Ehrlichia species closely related to Ehrlichia chaffeensis by sequence analyses of 16S ribosomal DNA in Boophilus microplus ticks from Tibet. J Clin Microbiol. 2002;40:3286–90. https://doi.org/10.1128/jcm.40.9.3286-3290.2002.
Marumoto K, Joncour G, Lamanda P, Inokuma H, Brouqui P. Detection of Anaplasma phagocytophilum and Ehrlichia sp HF strains in Ixodes ricinus ticks in Brittany, France. Clin Microbiol Infect. 2007;13:338–41. https://doi.org/10.1111/j.1469-0691.2006.01630.x.
Andersson MO, Radbea G, Frangoulidis D, Tomaso H, Rubel F, Nava S, et al. New records and host associations of the tick Ixodes apronophorus and the first detection of Ehrlichia sp HF in Romania. Parasitol Res. 2018;117:1285–9. https://doi.org/10.1007/s00436-018-5800-3.
Andersson MO, Tolf C, Tamba P, Stefanache M, Radbea G, Frangoulidis D, et al. Molecular survey of neglected bacterial pathogens reveals an abundant diversity of species and genotypes in ticks collected from animal hosts across Romania. Parasit Vectors. 2018;11:144. https://doi.org/10.1186/s13071-018-2756-1.
Hornok S, Boldogh SA, Takacs N, Juhasz A, Kontschan J, Foldi D, et al. Anaplasmataceae closely related to Ehrlichia chaffeensis and Neorickettsia helminthoeca from birds in Central Europe, Hungary. Anton Leeuw Int J G. 2020;113:1067–73. https://doi.org/10.1007/s10482-020-01415-4.
Santos-Silva MM, Beati L, Santos AS, De Sousa R, Nuncio MS, Melo P, et al. The hard-tick fauna of mainland Portugal (Acari: Ixodidae): an update on geographical distribution and known associations with hosts and pathogens. Exp Appl Acarol. 2011;55:85–121. https://doi.org/10.1007/s10493-011-9440-x.
Guglielmone AA, Nava S. Names for Ixodidae (Acari: Ixodoidea): valid, synonyms, incertae sedis, nomina dubia, nomina nuda, lapsus, incorrect and suppressed names-with notes on confusions and misidentifications. Zootaxa. 2014;3767:1–256.
Raoult D, Parola P. Rickettsial Diseases. 1st ed. Florida: CRC Press; 2007.
Acknowledgements
We gratefully acknowledge the help of the personnel of the Epidemiological Surveillance Program in Wildlife (Regional Government of Andalusia) in the collection of the samples and epidemiological information.
Funding
This study was partially funded by the Spanish Ministry of Science and Innovation (project LagoHealth; reference PID2019-111080RB-C21) and by the University of Córdoba (reference UCO-FEDER-1264967). The research was also supported by CIBER—Consorcio Centro de Investigación Biomédica en Red—(CB 2021), Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación and Unión Europea—NextGenerationEU, and is part of the TED2021-132599B-C22 project, funded by MCIN/AEI/10.13039/501100011033 and by the European Union NextGenerationEU/PRTR, and the Recovery, Transformation and Resilience Plan, funded by the European Union—NextGenerationEU. D. Jiménez-Martín holds a PhD contract granted by the Own Research Plan of the University of Cordoba. S. Castro-Scholten is supported by an FPU grant from the Spanish Ministry of Universities (FPU19/06026).
Author information
Authors and Affiliations
Contributions
SR and IGB wrote the original draft of the manuscript. SR, PM, DCT and IGB identified the research question and selected the methodology. SCS, DJM, CR, LCS, DCT and IGB conducted the field sampling. SR, PM and PD performed the experimental work. All the authors contributed to the critical review of the results and approved the final version of the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
No ethical approval was required since no animals were killed specifically for this study. The ticks that were analysed were collected from wild rabbits legally hunted in complete accordance with Andalusian and Spanish regulations.
Competing interests
The authors have no competing interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Remesar, S., Castro-Scholten, S., Morrondo, P. et al. Molecular detection of Ehrlichia spp. in ticks parasitizing wild lagomorphs from Spain: characterization of a novel Ehrlichia species. Parasites Vectors 15, 467 (2022). https://doi.org/10.1186/s13071-022-05600-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-022-05600-4