Abstract
Background
Ticks and tick-borne diseases constitute a real threat for the livestock industry, which is increasing in Angola. In addition, ticks are vectors of zoonoses of public health concern, and scarce information is available from this country. In an effort to contribute to the prevention of zoonotic infectious diseases affecting humans and animals, the molecular screening of certain tick-related microorganisms collected on cattle in Angola was performed under a ‘One Health’ scope.
Methods
Ticks collected from cattle in Cubal (Benguela Province, Angola) in July 2017 were analysed in pools using specific PCR assays for bacteria (Rickettsia, Anaplasmataceae, Borrelia, Coxiella and Spiroplasma) and protozoa (Theileria and Babesia) detection.
Results
A total of 124 tick specimens were grouped in 25 pools (two Amblyomma variegatum, three Hyalomma truncatum, 16 Rhipicephalus decoloratus, two Rhipicephalus duttoni, one Rhipicephalus evertsi mimeticus and one Rhipicephalus sp.). The amplified microorganisms were (pools): Rickettsia africae (two A. variegatum and one R. decoloratus), Rickettsia aeschlimannii (three H. truncatum), Ehrlichia spp. (six R. decoloratus), Coxiella spp. (all but H. truncatum), Francisella sp. (one H. truncatum), Spiroplasma sp. closely related to Spiroplasma ixodetis (three R. decoloratus), Babesia bigemina (two R. decoloratus) and Babesia spp. (two A. variegatum). The obtained nucleotide sequences from Ehrlichia spp., two Coxiella genotypes (from R. duttoni and Rhipicephalus sp.), Francisella sp. and Babesia spp. (from A. variegatum) reached low identities with known genetically characterized species.
Conclusions
This study demonstrates the circulation in Angola of the pathogen R. aeschlimannii and potential novel tick-related microorganisms belonging to Ehrlichia, Coxiella, Francisella, Spiroplasma and Babesia spp. and corroborates the presence of R. africae and B. bigemina. Our results should be considered in developing protocols for the management of fever of unknown origin and for veterinary practices. Further studies are required to evaluate the risk of tick-borne diseases in Angola.
Graphical Abstract

Similar content being viewed by others
Background
Epidemics and pandemics have been present throughout history, but the ongoing COVID-19 pandemic and recent epidemics have strengthened the importance of ‘One Health’ to prevent spillover events [1]. Human/animal health and the environment are interconnected, and factors such as globalization, climate change, changes in land use and population growth could trigger new zoonotic outbreaks [2]. Early detection and knowledge of potential zoonotic agents, including vector-borne microorganisms, is crucial to implementing containment measures and preventing related infectious diseases. Ticks are prominent vectors of zoonoses that pose a public health risk, such as Crimean-Congo haemorrhagic fever that is considered a ‘priority disease’ by the World Health Organization, due to their epidemic potential and/or lack of sufficient countermeasures [3, 4]. Thus, surveillance systems for vectors and their microorganisms are critically needed.
Zoonotic agents, often underdiagnosed due to lack of diagnostic resources, are a known major cause of disease in sub-Saharan Africa, and studies have revealed the need for improved protocols for fever of unknown origin (FUO) management [5]. Tick-borne relapsing fever, rickettsiosis and babesiosis have been reported from southern Africa [5, 6], but tick-borne diseases from Angola are hardly known. Ticks are of unquestionable veterinarian concern worldwide and constitute a real threat for the livestock industry, with a higher impact in poor countries [7]. Diseases such as heartwater (caused by Ehrlichia ruminantium) or theileriosis (caused by Theileria spp.) are endemic in sub-Saharan Africa [7, 8]. The Angolan livestock population is increasing (https://www.fao.org/faostat/en/#data/QCL), mainly based on cattle production, and the expansion of livestock industry is linked to the incidence of zoonosis [9]. Therefore, the molecular screening of selected microorganisms of public health concern in ticks infesting cattle during a week of July 2017 in the slaughterhouse of Cubal (Angola) is reported.
Methods
Ticks were collected from cattle in a slaughterhouse of Cubal (Benguela Province, Angola) from 1 to 8 July 2017, and preserved in 70% ethanol. Specimens were classified using a taxonomic key [10]. Selected individuals (at least two specimens from each morphologically classified species and those doubtful according to morphological features) were genetically characterized by polymerase chain reaction (PCR) of mitochondrial genes (Additional file 1: Table S1) using individual DNA. DNA was extracted from a leg of each specimen using two incubations of 20 min each with 100 µL of ammonium hydroxide 0.7 M at 100 and 90 °C, respectively [11]. Furthermore, tick halves were pooled (1–9 specimens) according to species and developmental stages. DNA from pools was extracted using a DNeasy Blood & Tissue kit (Qiagen), following the manufacturer’s recommendations with overnight lysis. Mitochondrial 16S rRNA PCRs were performed as controls of pool extractions (Additional file 1: Table S1). Bacteria (Rickettsia, Anaplasmataceae, Borrelia, Coxiella and Spiroplasma) and protozoa (Theileria and Babesia) were screened using specific PCR assays. Pan-bacterial 16S rRNA PCR was also performed (Additional file 1: Table S1).
The PCR amplicons obtained with the expected size were sequenced in forward and reverse senses. The nucleotide sequences were analysed using BioEdit v.7.2.6 software [12]. The consensus sequences produced were compared with those available in NCBI using BLAST [13], and submitted to GenBank when different. Clustal Omega [14] was used for multiple sequence alignment. Phylogenetic analyses were conducted with MEGA X [15] using maximum likelihood method including all sites. Confidence values for individual branches of the resulting trees were determined by bootstrap analysis (500 replicates).
Results
A total of 124 ticks (five nymphs, 28 males and 91 females) were collected and morphologically classified as six Amblyomma variegatum, six Hyalomma truncatum, 107 Rhipicephalus decoloratus and five Rhipicephalus spp. Whenever performed, genetic characterization confirmed morphological identification, and also allowed the identification of three Rhipicephalus duttoni and one Rhipicephalus evertsi mimeticus (Tables 1, 2) among those Rhipicephalus spp.
Twenty-five pools (two A. variegatum, three H. truncatum, 16 R. decoloratus, two R. duttoni, one R. evertsi mimeticus and one Rhipicephalus sp.) were screened for microorganisms.
Rickettsia spp. was found in 6/25 pools. According to ompA, Rickettsia africae was detected in two A. variegatum pools and one R. decoloratus pool, and R. aeschlimannii in three H. truncatum pools (Table 2). Ehrlichia spp. was found in 6/25 pools of female R. decoloratus. Analysis of groESL, gltA and 16S rRNA amplicons revealed the highest identities with unclassified Ehrlichia (Table 2, Fig. 1a, b) and showed less than 93.5%, 87.6% and 99.2% identity, respectively, with validated species. Other Anaplasmataceae, Borrelia spp. (relapsing fever or Lyme groups) or Coxiella burnetii were not detected. Nevertheless, Coxiella spp. were found in all but H. truncatum pools. For H. truncatum, rpoB sequences showed inconclusive data, whereas groEL and universal 16S rRNA sequences showed the highest similarity (< 97% and 99.6%, respectively) with Francisella sp. in one pool. This 16S rRNA amplicon showed 99.8% identity (92% query cover) with a Francisella endosymbiont of H. truncatum JF290387 (Table 2). For the remaining tick species, different Coxiella genotypes were found. All but two were identical or closely related to public sequences. Genotypes detected in R. duttoni and Rhipicephalus sp. did not reach > 98.3% identity with Coxiella (Table 2, Fig. 1c). Spiroplasma sp. was amplified from three R. decoloratus male pools (Table 2). According to rpoB, this genotype was closely related to Spiroplasma ixodetis and related strains of hard ticks (Fig. 1d).
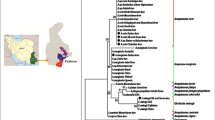
Phylogenetic analysis of the microorganisms detected in this study from ticks collected from cattle in Angola (marked with diamonds). The maximum likelihood trees were obtained using the general time reversible model, a discrete gamma distribution and a proportion of invariable sites (GTR + G + I), with nucleotide substitution selected according to the Akaike information criterion implemented in Mega X. The trees are drawn to scale, with branch lengths measured in the number of substitutions per site. Numbers (> 60%) shown at the nodes correspond to bootstrapped percentages (for 500 repetitions). The GenBank accession numbers of the sequences used in these analyses are shown in brackets. a Ehrlichia phylogeny was based on 23 partial 16S rRNA gene sequences with a total of 1373 positions in the final dataset. Candidatus Neoehrlichia mikurensis was used as an outgroup. b Ehrlichia phylogeny was based on 22 partial groESL gene sequences with a total of 1232 positions in the final dataset. Candidatus Neoehrlichia mikurensis was used as an outgroup. c Coxiella-like phylogeny was based on 51 partial rpoB and groEL concatenated sequences with a total of 1055 positions in the final dataset. Rickettsiella sp. was used as an outgroup. d Phylogeny of Spiroplasma spp. found in ticks based on 18 partial rpoB sequences with a total of 588 positions in the final dataset. e Phylogeny of Babesia species based on 18S rRNA analysis. The analysis involved 40 nucleotide sequences and a total of 481 positions in the final dataset. Plasmodium falciparum was used as outgroup
Babesia bigemina was identified in two R. decoloratus female pools, and Babesia sp. was detected in two A. variegatum pools, according to 18S rRNA, ITS-1 and ITS-2 analysis (Table 2, Fig. 1e).
Discussion
This study reports the detection of well-known pathogens R. africae, R. aeschlimannii and B. bigemina, and scarce characterized Ehrlichia, Coxiella, Francisella, Spiroplasma and Babesia species with unknown pathogenicity in ticks from cattle in Angola.
Our results confirm the circulation of R. africae and demonstrate the circulation of R. aeschlimannii in Angola. Although R. aeschlimannii human infection had been reported from South Africa and H. truncatum had been suggested as a vector [16, 17], this pathogen had not been previously found in Angola. African tick-bite fever (ATBF) is endemic in sub-Saharan Africa, but no cases from Angola have been reported [5, 6]. The agent, R. africae, has been recently reported from A. variegatum in this area [18]. This study confirms the circulation of R. africae in the recognized vector, suggesting that ATBF cases could be unreported or misdiagnosed. The presence of R. africae in R. decoloratus is known, but their role as a vector should be investigated [6, 19]. Moreover, our finding in fed ticks could be due to blood meal or co-feeding.
Only six Ehrlichia species are currently recognized, and all but one are responsible for human and/or animal ehrlichiosis [20]. Human ehrlichiosis has been reported from southern Africa, where heartwater (a disease of domestic and wild ruminants caused by E. ruminantium) is endemic [6, 8]. Moreover, ‘Candidatus’ have been proposed, and Ehrlichia genotypes have been partially characterized. Further studies are needed to determine their taxonomic status and pathogenic potential. Herein, a novel Ehrlichia genotype has been detected in six R. decoloratus pools.
Tick diet based on blood is unbalanced, and endosymbionts (e.g. Coxiella-like, Francisella-like) provide essential nutrients for ticks [21]. Although virulence genes identified in their pathogenic related species, C. burnetii and Francisella tularensis, could be absent or non-functional in symbionts, Coxiella-like has been considered a pathogen [21,22,23]. Herein, Coxiella-like was detected in all but H. truncatum pools, and potential novel Coxiella genotypes were detected in R. duttoni and Rhipicephalus sp. The remaining amplicons showed sequences identical or closely related to Coxiella-like previously amplified in the same tick species. Francisella sp. was detected in 1/3 H. truncatum pools. The sequence was genetically related to a Francisella sp. endosymbiont amplicon previously detected in this tick species.
Spiroplasma spp. have been found in several hard tick species, and the role of this genus as pathogen has been suggested [24]. Herein, Spiroplasma sp. closely related to S. ixodetis was detected in 3/16 R. decoloratus pools. Spiroplasma sp. was previously detected in this species according to a short rpoB sequence (Table 2), and this study provides a wider genetic identification.
Babesia bigemina, responsible for babesiosis, is prevalent in Angolan cattle [25]. Our study demonstrates its presence in R. decoloratus (competent vector) in Angola. Moreover, a potential novel Babesia species is circulating in Angolan A. variegatum.
Ehrlichia ruminantium, Anaplasma and Theileria spp. have been reported from Angolan cattle; the latter has been also detected in one A. variegatum specimen [8, 18, 25, 26]. Herein, the failure to detect these expected tick-borne microorganisms could be due to the low number of ticks and species analysed, the short period of time for tick collection and/or the host origin.
Conclusions
This study highlights the importance of ticks in public health in the studied area, and these results should be considered in developing protocols for the management of patients with FUO and for veterinary practices in Angola. Nevertheless, this is only the tip of the iceberg, and more ticks belonging to more species, hosts, from wider areas, etc., as well as broader screening of microorganisms, including viruses (not analysed in this study due to the sample preservation method available), are required to evaluate the risk of tick-borne diseases in Angola.
Availability of data and materials
Novel sequences of this study were deposited on GenBank under accession numbers OK481091-OK481100, OK481107-OK481113, OK491113-OK491116, OK482869-OK482874 and OK514711-OK514725.
Abbreviations
- ATBF:
-
African tick-bite fever
- BLAST:
-
Basic Local Alignment Search Tool
- COI:
-
Cytochrome oxidase subunit 1
- COVID-19:
-
Coronavirus disease 2019
- DNA:
-
Deoxyribonucleic acid
- FUO:
-
Fever of unknown origin
- GenBank:
-
NIH (National Institutes of Health) genetic sequence database
- gltA :
-
Citrate synthase
- groEL:
-
Chaperonin groEL
- groESL:
-
Chaperonin GroEL/GroES complex
- ITS:
-
Internal transcribed spacer
- NCBI:
-
National Center for Biotechnology Information
- ompA :
-
Outer membrane protein A
- PCR:
-
Polymerase chain reaction
- rpoB :
-
RNA polymerase beta subunit
- rRNA:
-
Ribosomal ribonucleic acid
- sp./spp.:
-
Species
References
Pitlik SD. COVID-19 compared to other pandemic diseases. Rambam Maimonides Med J. 2020;11: e0027. https://doi.org/10.5041/RMMJ.10418.
Otu A, Effa E, Meseko C, Cadmus S, Ochu C, Athingo R, et al. Africa needs to prioritize One Health approaches that focus on the environment, animal health and human health. Nat Med. 2021;27:943–6. https://doi.org/10.1038/s41591-021-01375-w.
World Health Organization. Prioritizing Diseases for Research and Development in Emergency Contexts. https://www.who.int/activities/prioritizing-diseases-for-research-and-development-in-emergency-contexts. Accessed 1 Feb 2022.
Portillo A, Palomar AM, Santibáñez P, Oteo JA. Epidemiological aspects of Crimean-Congo hemorrhagic fever in Western Europe: what about the future? Microorganisms. 2021;9:649. https://doi.org/10.3390/microorganisms9030649.
Maze MJ, Bassat Q, Feasey NA, Mandomando I, Musicha P, Crump JA. The epidemiology of febrile illness in sub-Saharan Africa: implications for diagnosis and management. Clin Microbiol Infect. 2018;24:808–14. https://doi.org/10.1016/j.cmi.2018.02.011.
Chitanga S, Gaff H, Mukaratirwa S. Tick-borne pathogens of potential zoonotic importance in the southern African Region. J S Afr Vet Assoc. 2014;85:1084. https://doi.org/10.4102/jsava.v85i1.1084.
Minjauw B, McLeod A. Tick-borne diseases and poverty. The impact of ticks and tickborne diseases on the livelihood of small-scale and marginal livestock owners in India and eastern and southern Africa. Research report. UK; DFID Animal Health Programme, Centre for Tropical Veterinary Medicine, University of Edinburgh; 2003
Gomes AF. The tick vectors of cowdriosis in Angola. Rev Elev Med Vet Pays Trop. 1993;46:237–43.
Rohr JR, Barrett CB, Civitello DJ, Craft ME, Delius B, DeLeo GA, et al. Emerging human infectious diseases and the links to global food production. Nat Sustain. 2019;2:445–56. https://doi.org/10.1038/s41893-019-0293-3.
Walker AR, Bouattour A, Camicas JL, Estrada-Peña A, Horak IG, Latif AA, et al. Ticks of domestic animals in Africa: a guide to identification of species. Biosci Rep. 2003;201.
Portillo A, Santos AS, Santibáñez S, Pérez-Martínez L, Blanco JR, Ibarra V, et al. Detection of a non-pathogenic variant of Anaplasma phagocytophilum in Ixodes ricinus from La Rioja, Spain. Ann NY Acad Sci. 2005;1063:333–6. https://doi.org/10.1196/annals.1355.053.
Hall TA. BioEdit: A User-friendly Biological Sequences Alignment Editor and Analysis Program for Windows 95/98/NT. Nucleic Acids Symposium Series No. 41. Oxford: Oxford University Press; 1999. p. 95–98
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. https://doi.org/10.1016/S0022-2836(05)80360-2.
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47:W636–41. https://doi.org/10.1093/nar/gkz268.
Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35:1547–9. https://doi.org/10.1093/molbev/msy096.
Pretorius AM, Birtles RJ. Rickettsia aeschlimannii: a new pathogenic spotted fever group rickettsia, South Africa. Emerg Infect Dis. 2002;8:874. https://doi.org/10.3201/eid0808.020199.
Mediannikov O, Diatta G, Fenollar F, Sokhna C, Trape JF, Raoult D. Tick-borne rickettsioses, neglected emerging diseases in rural Senegal. PLoS Negl Trop Dis. 2010;4: e821. https://doi.org/10.1371/journal.pntd.0000821.
Barradas PF, Mesquita JR, Ferreira P, Gärtner F, Carvalho M, Inácio E, et al. Molecular identification and characterization of Rickettsia spp. and other tick-borne pathogens in cattle and their ticks from Huambo, Angola. Ticks Tick Borne Dis. 2021;12:101583. https://doi.org/10.1016/j.ttbdis.2020.101583.
Portillo A, Pérez-Martínez L, Santibáñez S, Blanco JR, Ibarra V, Oteo JA. Detection of Rickettsia africae in Rhipicephalus (Boophilus) decoloratus ticks from the Republic of Botswana, South Africa. Am J Trop Med Hyg. 2007;77:376–7.
Saito TB, Walker DH. Ehrlichioses: an important one health opportunity. Vet Sci. 2016;3:20. https://doi.org/10.3390/vetsci3030020.
Angelakis E, Mediannikov O, Jos SL, Berenger JM, Parola P, Raoult D. Candidatus Coxiella massiliensis infection. Emerg Infect Dis. 2016;22:285–8. https://doi.org/10.3201/eid2202.150106.
Buysse M, Duron O. Evidence that microbes identified as tick-borne pathogens are nutritional endosymbionts. Cell. 2021;184:2259–60. https://doi.org/10.1016/j.cell.2021.03.053.
Buysse M, Floriano AM, Gottlieb Y, Nardi T, Comandatore F, Olivieri E. A dual endosymbiosis supports nutritional adaptation to hematophagy in the invasive tick Hyalomma marginatum. Elife. 2021;10: e72747. https://doi.org/10.7554/eLife.72747.
Palomar AM, Premchand-Branker S, Alberdi P, Belova OA, Moniuszko-Malinowska A, Kahl O, et al. Isolation of known and potentially pathogenic tick-borne microorganisms from European ixodid ticks using tick cell lines. Ticks Tick Borne Dis. 2019;10:628–38. https://doi.org/10.1016/j.ttbdis.2019.02.008.
Sili G, Byaruhanga C, Horak I, Steyn H, Chaisi M, Oosthuizen MC, et al. Ticks and tick-borne pathogens infecting livestock and dogs in Tchicala-Tcholoanga, Huambo Province, Angola. Parasitol Res. 2021;120:1097–102. https://doi.org/10.1007/s00436-020-07009-3.
Kubelová M, Mazancová J, Široký P. Theileria, Babesia, and Anaplasma detected by PCR in ruminant herds at Bie Province, Angola. Parasite. 2012;19:417–22. https://doi.org/10.1051/parasite/2012194417.
Acknowledgements
Partial results of this study were presented at the SEIMC [Spanish Society of Clinical Microbiology and Infectious Diseases] XXIII National Congress (Madrid, Spain, 2019).
Funding
This work has been partially funded by European Regional Development Funds (FEDER).
Author information
Authors and Affiliations
Contributions
AMP, IM, CB, FS and JAO conceived and designed the study. AMP, AP and JAO designed the methodology for the microorganism screening. IM, CB, FS and MM carried out the field work. AMP identified ticks, processed samples, and performed PCR assays, sequencing analysis and phylogenies. AMP, IM, AP, FS and JAO analysed the data. AMP and JAO drafted the manuscript. All authors reviewed the original manuscript and agreed to the final version. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
PCR primer pairs and conditions used in this study.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Palomar, A.M., Molina, I., Bocanegra, C. et al. Old zoonotic agents and novel variants of tick-borne microorganisms from Benguela (Angola), July 2017. Parasites Vectors 15, 140 (2022). https://doi.org/10.1186/s13071-022-05238-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-022-05238-2