Abstract
Background
Our study aimed to assess the diversity of the species of Anaplasmataceae in Senegal that infect animals and ticks in three areas: near Keur Momar Sarr (northern region), Dielmo and Diop (Sine Saloum, central region of Senegal), and in Casamance (southern region of Senegal).
Methods
A total of 204 ticks and 433 blood samples were collected from ruminants, horses, donkeys and dogs. Ticks were identified morphologically and by molecular characterization targeting the 12S rRNA gene. Molecular characterization of species of Anaplasmataceae infecting Senegalese ticks and animals was conducted using the 23S rRNA, 16S rRNA, rpoB and groEL genes.
Results
Ticks were identified as Rhipicephalus evertsi evertsi (84.3%), Hyalomma rufipes (8.3%), Hyalomma impeltatum (4.9%), R. bursa (1.5%) and R. muhsamae (0.9%). The overall prevalence of Anaplasmataceae infection in ticks was 0.9%, whereas 41.1% of the sampled animals were found infected by one of the species belonging to this family. We identified the pathogen Anaplasma ovis in 55.9% of sheep, A. marginale and A. centrale in 19.4% and 8.1%, respectively, of cattle, as well as a putative new species of Anaplasmataceae. Two Anaplasma species commonly infecting ruminants were identified. Anaplasma cf. platys, closely related to A. platys was identified in 19.8% of sheep, 27.7% of goats and 22.6% of cattle, whereas a putative new species, named here provisionally “Candidatus Anaplasma africae”, was identified in 3.7% of sheep, 10.3% of goats and 8.1% of cattle. Ehrlichia canis and Anaplasma platys were identified only from dogs sampled in the Keur Momar Sarr area. Ehrlichia canis was identified in 18.8% of dogs and two R. e. evertsi ticks removed from the same sheep. Anaplasma platys was identified in 15.6% of dogs. Neither of the dogs sampled from Casamance region nor the horses and donkeys sampled from Keur Momar Sarr area were found infected by an Anaplasmataceae species.
Conclusions
This study presents a summary of Anaplasmataceae species that infect animals and ticks in three areas from the northern, central and southern regions of Senegal. To our knowledge, our findings demonstrate for the first time the presence of multiple Anaplasmataceae species that infect ticks and domestic animals in Senegal. We recorded two potentially new species commonly infecting ruminants named here provisionally as Anaplasma cf. platys and “Candidatus Anaplasma africae”. However, E. canis was the only species identified and amplified from ticks. None of the other Anaplasmataceae species identified in animals were identified in the tick species collected from animals.
Similar content being viewed by others
Background
A member of the order Rickettsiales, the family Anaplasmataceae contains the zoonotic intracellular alpha-proteobacteria of the genera Anaplasma, Ehrlichia, Neoehrlichia and Neorickettsia [1]. These vector-borne bacteria are transmitted mainly by ixodid ticks (Anaplasma, Ehrlichia and Neoehrlichia) whereas Neorickettsia are intracellular endosymbionts of a diverse group of the Digenea (Platyhelminthes: Trematoda) [2]. In ticks, transmission of Anaplasma and Ehrlichia species occurs transtadially but not transovarially; therefore, every tick generation must obtain infection by feeding on reservoir hosts [3]. Anaplasma and Ehrlichia are able to cause a persistent infection in the vertebrate hosts, which allows them to be reservoirs of the infection [4, 5]. The nature of the infection cycle and the virulence of different strains of Anaplasma and Ehrlichia depend on the susceptibility of the infected vertebrate hosts and the availability and abundance of ixodid tick vectors largely interconnected in an epidemiological network [6, 7]. The persistent infection induced by Anaplasma or Ehrlichia can cause death in animals due to co-infection by Staphylococcus aureus or Mannheimia/Bibersteinia, “pasteurellosis” and other opportunistic diseases [8]. Animals are variably susceptible to the different strains of Anaplasma and Ehrlichia. For example, the American Anaplasma phagocytophilum strain human-active (Ap-ha) and variant 1 (Ap-v1) [9] seem to be less pathogenic to animals and fail to induce disease or marked bacteremia [10]. However, the European A. phagocytophilum strains are pathogens for cattle, sheep, goats, dogs and cats [11]. Bovine anaplasmosis caused by A. marginale is a worldwide reported infection. It results in the development of mild to severe anemia [12]. Anaplasma marginale and Babesia spp. together are responsible for economic losses reaching 22 and 57 million USD in Australia and India, respectively [12, 13]. Anaplasma ovis is a neglected agent of sheep and goat anaplasmosis due to the usually subclinical course of the disease [14]. Research in the last decade has further elucidated the nature of the syndrome caused by anaplasmosis in the infected host, the importance of animals as a reservoir of this bacteria, and the zoonotic potential of some Anaplasma spp. [5, 15, 16].
In Africa, the prevalence of Anaplasmataceae infection and extent to which livestock productivity has been affected remain poorly understood. These bacteria were recorded in many countries in southern Africa but few studies have been conducted in West Africa [17]. To better understand the epidemiological significance of the Anaplasmataceae infection in animals, it is necessary to include in the analyses multiple samples from different mammalian hosts and vectors.
The current reported statistics about livestock numbers, distribution and economic importance are difficult to evaluate in Senegal. Our main objective was to provide updated information about the local spread and epidemiology of infectious diseases in animals and ticks in three areas near Keur Momar Sarr (northern region of Senegal), Dielmo and Diop (Sine Saloum, central region of Senegal) and Casamance (southern region of Senegal). The aim of this study was to provide a detailed overview regarding the presence and the prevalence of Anaplasmataceae species infecting and currently circulating in and between cattle, sheep, goats, horses, donkeys, dogs and ticks in this region, to evaluate the genetic diversity of these bacteria and, finally, to carry out their phylogenetic characterization.
Methods
Period, study areas and collection of ticks and blood samples
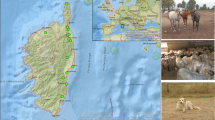
All animals and ticks were sampled and collected in June 2013 and June 2014 from three Senegalese areas (Fig. 1): (i) the northern region near Keur Momar Sarr (15°55′0.0012″N, 15°58′0.0012″W): Gankette Balla (15°58′50.6”N, 15°55′42.6”W), Loboudou (15°57′10.98″N, 15°55′11.8668″W) and Ndimb (16°2′56.958″N, 16°0′10.5876″W); (ii) Sine Saloum (central region of Senegal near Gambia): Dielmo (13°43′0.0012″N, 16°23′60″W) and Diop (13°40′59.9988″N, 16°21′59.976″W); and (iii) Casamance (southern region): Oussouye (12°29′13.8768″N, 16°32′52.8288″W). Animals were examined with the assistance of their owners. Ticks and blood samples were collected by a veterinarian. Overall, in 2013, 47 blood samples from 47 cattle were sampled in Dielmo and Diop, and 78 dog blood samples were collected in Casamance. In 2014, other EDTA blood samples and ticks were also collected from 136 sheep, 29 goats, 15 cattle, 64 horses, 29 donkeys and 64 dogs in the Keur Momar Sarr area (Table 1). Two hundred four adult ticks were manually collected from animals. Blood samples and ticks (stored in 70% ethanol) were transported to the laboratory of the IHU Méditerranée Infection, Marseille (France). Upon arrival, all blood samples were stored at − 80 °C. Ticks in ethanol were stored at room temperature.
Ticks were identified morphologically under a binocular microscope. Ticks were classified by family, genus and species using available taxonomic keys and morphometric tables [18,19,20]. In addition, to confirm the morphological identification, three ticks from each tick species and all ticks that were not identified or only identified at the family level, including engorged females and damaged ticks, were subjected to a molecular identification using primers targeting the mitochondrial 12S rDNA as described previously [21].
DNA extraction
DNA extraction was performed on a Bio Robot EZ1 (Qiagen, Courtaboeuf, France) using a commercial EZ1 DNA Tissue Kit (Qiagen) according to the manufacturer’s instructions. DNA was extracted from 200 µl of blood from all the animal samples. Ticks were recovered from ethanol, rinsed with distilled water and dried on sterile filter paper in a laminar-flow hood. Each tick was cut in half lengthways (the blades were discarded after each tick was cut). DNA was individually extracted from one-half, and the remaining tick halves were frozen at − 80 °C for subsequent studies as previously described [22].
PCR amplification
DNA samples from the selected tick species were subjected to PCR amplification using primers targeting the 360-bp long fragment of the mitochondrial 12S rDNA gene. In order to investigate the presence of Anaplasmataceae in Senegalese ticks and in domestic animal blood samples, DNA samples were initially screened by a qPCR targeting the 23S rRNA gene. This qPCR determined that most bacteria belonged to the family Anaplasmataceae [23]. Then, all positive samples were subjected to a conventional PCR using primers that amplify a 485-bp long fragment of the 23S rRNA gene, as previously described [24]. In order to mine deeper into the identity of selected Anaplasmataceae species in domestic animals or ticks, one or more DNA samples representative of each Anaplasmataceae species previously identified by the 23S rRNA gene were amplified using primers specifically targeting the 16S rRNA gene for the family Anaplasmataceae (828 bp), the RNA polymerase subunit beta (rpoB) gene for the genus Anaplasma (525 bp), and specific primers targeting the heat-shock protein (groEL) gene for the genus Ehrlichia (590 bp) (Table 2).
PCR amplifications were performed as described previously [23, 24]. The real-time PCR assays were performed on a CFX96 Touch detection system (Bio-Rad, Marnes-la-Coquette, France) using Takyon Master Mix under the conditions suggested by the manufacturer. The conventional PCRs were performed in automated DNA thermal cyclers (GeneAmp PCR Systems, Applied Biosystems, Courtaboeuf, France). The amplification reactions were performed under the following conditions: an initial denaturation step at 95 °C for 15 min, followed by 40 cycles of 1 min denaturation at 95 °C, 1 min annealing at a corresponding temperature (Table 2) and 1 min extension at 72 °C. A final extension cycle at 72 °C for 7 min was performed, and the reactions were cooled to 15 °C. Distilled water and DNA of A. phagocytophilum obtained from HL60 infected cell line maintained in our laboratory, and Ehrlichia canis obtained from old DNA extracted from infected dogʼs blood sampled in Algeria from our previous study [24] were used in each test as negative and positive controls, respectively. The amplification products were visualized on 1.5% agarose gels stained with ethidium bromide and examined by UV transillumination. A DNA molecular weight marker (marker VI, Boehringer, Mannheim, Germany) was used to estimate the sizes of the products.
Sequencing and phylogenetic analyses
Sequencing analyses were performed on an Applied Biosystems 3130xl Genetic Analyzer (Thermo Fisher Scientific, Les Ulis, France) using the DNA sequencing Big Dye Terminator Kit (Perkin-Elmer, Hamburg, Germany) as described by the manufacturer. The obtained sequences were assembled using ChromasPro v.1.7 software (Technelysium Pty Ltd., Tewantin, Australia). The sequences of primers were removed and the newly generated sequences were aligned with other tick or Anaplasmataceae species sequences available on GenBank using CLUSTAL W implemented in BioEdit v.3 [25]. The sequences of 12S rDNA from ticks and the sequences of the 23S rRNA, rpoB and groEL genes were first aligned individually; gaps and missing data were eliminated. Then, the sequence alignments of the 23S rRNA, 16S rRNA with rpoB genes for Anaplasmataceae species, and 23S rRNA, 16S rRNA with groEL genes for Ehrlichia species were concatenated for phylogenetic tree construction. Phylogenetic relationships and molecular evolution were inferred using the maximum likelihood method implemented in MEGA7 [26], with the complete deletion option, based on the Hasegawa–Kishino–Yano model. A discrete gamma distribution was used to model evolutionary rate differences among sites. Initial trees for the heuristic search were obtained automatically by applying the Neighbor-Join and BioNJ algorithms to a matrix of pair wise distances estimated using the maximum composite likelihood approach. Statistical support for internal branches of the trees was evaluated by bootstrapping with 1000 iterations.
Results
Ticks morphological and molecular identification
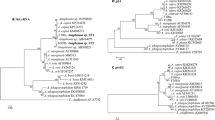
All ticks were collected in the Keur Momar Sarr area. A total of 204 ticks were collected; of these, 64.2% (131) were male and 35.8% (73) were female. One hundred thirty-nine ticks were removed from 26 horses, 46 ticks from 15 sheep, 9 ticks from 3 cattle, 6 ticks from 2 donkeys and 2 ticks from 2 dogs (Table 1). One to 16 ticks were collected from each animal. One hundred seventy-seven ticks were morphologically identified as follows: 154 Rhipicephalus evertsi evertsi (75.5%), 14 Hyalomma rufipes (6.9%) and 9 Hyalomma impeltatum (4.4%). Twenty-seven ticks were not identified at species level. These ticks included fully-engorged and damaged female ticks (n = 25) and two ticks removed from two dogs identified at the Rhipicephalus sp. level. These ticks were subject to molecular characterization. In addition to ticks not identified morphologically, at least three samples of R. e. evertsi, Hy. impeltatum, and Hy. rufipes identified morphologically were subjected to further molecular identification. Finally, 47 ticks were used for the 12S rRNA gene amplification. Each sequence generated from each amplicon and belonging to the same species were aligned individually using CLUSTAL W; then, gaps and single-nucleotide polymorphisms (SNP) were corrected. The 25 previously unidentified engorged females were identified as follows: 18 Rhipicephalus evertsi evertsi, 1 Hy. impeltatum, 3 Hy. rufipes and 3 R. bursa. The two ticks taken from dogs were identified as R. muhsamae. From the 20 remaining ticks identified morphologically that were subjected to molecular characterization, we did not find any discordance between molecular and morphological identification. The sequences of R. e. evertsi showed 98% identity with R. e. evertsi collected in Zimbabwe (GenBank: AF150052) and Zambia (GenBank: DQ901291–DQ849229). The sequences of Hy. impeltatum showed 99% identity with Hy. impeltatum collected in Niger (GenBank: KX132904). Hyalomma rufipes showed 100% identity with the sequences of Hy. rufipes collected in France (GenBank: KX000618–KX000610) and Italy (GenBank: KC817342). The sequences of R. bursa showed 99% identity with the sequences of R. bursa collected in Italy (GenBank: AM410572, KC243833, KC243834 and KU512950) and Spain (GenBank: AF150053). Finally, the sequences of R. muhsamae showed 100% identity with R. muhsamae collected in Nigeria (GenBank: KC243829). The phylogenetic tree comparing the sequences of 12S rRNA gene amplified from our ticks to other sequences of the same gene available in GenBank is presented in Fig. 2.
Phylogenetic tree showing the position of R. evertsi evertsi, Hy. rufipes, Hy. impeltatum, R. bursa and R. muhsamae compared to other tick species. Evolutionary analyses were conducted using MEGA7 [26]. The sequences of the 12S rDNA amplified in this study with other 12S rDNA tick sequences available on GenBank were aligned using CLUSTAL W implemented on BioEdit v.3 [25] (there were 262 positions in the final dataset). The evolutionary history was inferred by using the maximum likelihood method based on the Hasegawa–Kishino–Yano model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log likelihood value. Statistical support for internal branches of the trees was evaluated by bootstrapping with 1000 iterations. A discrete gamma distribution was used to model evolutionary rate differences among sites [2 categories (+G, parameter = 0.4726)]. The analysis involved 52 nucleotide sequences. All positions containing gaps and missing data were excluded. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The scale-bar represents a 5% nucleotide sequence divergence
All tick species collected are listed in Table 1. Rhipicephalus evertsi evertsi were the most commonly collected ticks in all animals (172, 84.3%) except from dogs. Hyalomma rufipes (17, 8.3%) were collected from four cattle and 13 horses. Hyalomma impeltatum (10, 4.9%) and R. bursa (3, 1.5%) were both collected from three different horses. Finally, R. muhsamae (2, 0.9%) were removed only from dogs (Table 1). Occurrence of more than one tick species on a single animal was observed: 2 cattle were found infested with R. e. evertsi and Hy. rufipes; co-occurrence of R. e. evertsi with Hy. rufipes, and R. e. evertsi with R. bursa was observed in 5 and 3 horses, respectively; and one horse was found infested by three tick species, R. e. evertsi, Hy. rufipes and Hy. impeltatum.
Anaplasmataceae species screening and sequencing
All results are summarized in Table 2. The 637 DNA samples extracted from ticks and animal blood were first screened using qPCR targeting the 23S RNA gene of the Anaplasmataceae. In ticks, only two Rh. e. evertsi (0.9%) collected from the same sheep were found positive. In animals, 108/136 sheep (79.4%), 11/29 goats (37.9%), 38/62 cattle (61.3%) and 22/142 dogs (15.5%) were found positive. Sheep, goats and dogs positive for Anaplasmataceae in the qPCR were all from Keur Momar Sarr. For cattle, 29 positives were from the Sine Saloum in the central region of Senegal (29/47; 61.7%) and eight were from the Keur Momar Sarr in the northern region of Senegal (8/15; 53.3%).
Using primers that amplify a 485-bp long fragment of the 23S rRNA gene, all the Anaplasmataceae-positive samples identified in the qPCR were amplified and sequenced. The sequences generated from each amplicon belonging to the same species were aligned individually using CLUSTAL W; then, gaps and SNP were corrected. Ehrlichia canis was amplified from 2 R. e. evertsi and showed 99% identity with E. canis strain Jack (GenBank: NR076375). Anaplasma ovis was identified exclusively in sheep. The sequences showed 100% with A. ovis genotype KMND Niayes 14 (GenBank: KM021411). Anaplasma marginale was identified from cattle. The sequences showed 100% identity with A. marginale strain Dawn (GenBank: CP006847), Gypsy Plains (GenBank: CP006846), Florida (GenBank: NR076579) and 99% identity with A. centrale strain Israel (GenBank: NR076686). Anaplasma centrale was also identified in cattle blood samples (9.4%) and showed 100% identity with A. centrale strain Israel (GenBank: NR076686) and 99% identity with A. marginale strain Dawn (GenBank: CP006847), Gypsy Plains (GenBank: CP006846) and Florida (GenBank: NR076579). From cattle sampled in the Sine Saloum, we obtained sequences of poor quality in 11/47 cattle. BLAST analysis showed that these sequences had the same homology (98–99%) and belonged either to A. marginale or A. centrale. We removed these poor sequences from further analysis. From sheep, cattle and goats, identical sequences were obtained. They showed 99% identity with A. platys stain ChieGuy88 (GenBank: KM021414) reported from French Guiana, A. platys strain Dog Gard1 (GenBank: KM021412) reported from France and A. platys strain ChieCal05 (GenBank: KM021425) reported from New Caledonia. Finally, from sheep, cattle and goats, another potentially new species was identified. The obtained sequences of this species were identical to each other and showed 93% identity with A. marginale strain Dawn (GenBank: CP006847), Gypsy Plains (GenBank: CP006846) and Florida (GenBank: NR076579), and 92% identity with A. centrale strain Israel (GenBank: NR076686) (Table 2).
From dogs, A. platys were found to infect 10/64 (15.6%) of dogs. The sequences were identical to each other and showed 99% homology with A. platys amplified from sheep, cattle and goats in the present study and 99% identity with A. platys stain ChieGuy88 (GenBank: KM021414), A. platys strain Dog Gard1 (GenBank: KM021412) and A. platys strain ChieCal05 (GenBank: KM021425). In addition, 12/64 (18.8%) of dogs were also infected with Ehrlichia canis that showed 100% with the sequences of E. canis amplified from the two R. e. evertsi in the present study and 99% identity with E. canis strain Jack (GenBank: NR076375).
For further molecular identification targeting other genes for the family Anaplasmataceae, the two R. e. evertsi that harbored E. canis and for each different identified Anaplasmataceae species amplified by the primers targeting the 23S rRNA gene, three randomly selected samples were chosen and the DNA was used to amplify a 828-bp long fragment of the Anaplasmataceae family 16S rRNA, an Anaplasma spp. 525-bp long fragment of the RNA polymerase subunit beta (rpoB) gene and an Ehrlichia spp. 590-bp long fragment of the heat-shock protein (groEL) gene. The amplification followed by sequencing result showed that A. ovis 16S rRNA sequences amplified from sheep had 99% identity with A. ovis reported worldwide, whereas the rpoB sequences showed 100% identity with A. ovis strain RhburBas11 (GenBank: KX155495) reported from France and KMND Niayes 14 (GenBank: KX155494) reported from Senegal. Anaplasma marginale 16S rRNA sequences amplified from cattle showed 99% identity with multiple sequences of A. marginale reported from Uganda and with A. marginale strains Dawn (GenBank: CP006847) and Gypsy Plains (GenBank: CP006846). The rpoB sequences of this A. marginale strain showed 99% identity with A. marginale strain Dawn (GenBank: CP006847), Gypsy Plains (GenBank: CP006846) and Florida (GenBank: CP001079). The 16S rRNA sequences of A. centrale showed 99% of identity with A. centrale, A. ovis and A. marginale reported worldwide. The rpoB gene sequence of A. centrale has 99% identity with A. centrale strain Israel (GenBank: CP001759) and 88% identity with A. marginale strain Dawn (GenBank: CP006847). The16S rRNA and rpoB sequences of A. platys amplified from ruminant were amplified from three samples each taken from sheep, goats and cattle. For each gene, the sequences were identical to each other and showed for the 16S rRNA 99% identity with multiple uncultured Anaplasma sp., A. platys (GenBank: KY114935-EF139459), A. platys isolate A.pl. #87 (GenBank: JQ396431), “Candidatus Anaplasma camelii” clone Camel_38 (GenBank: KF843827) and 99% identity with A. phagocytophilum reported worldwide. The rpoB sequences showed 93% identity with A. platys strain Dog Gard1 (GenBank: KX155493) and 89% with A. phagocytophilum strain Norway variant2 (GenBank: CP015376). Sequence analysis of the three genes used in the present study revealed that this species is distinct from A. platys (99% for 23S rRNA, 99% for 16S rRNA and 93% for rpoB). Due to the absence of additional data on this Anaplasma sp. and the genetic relatedness to A. platys, we refer to this genotype here as Anaplasma cf. platys. A phylogenetic tree based on the concatenated 23S rRNA, 16S rRNA and rpoB genes showed that Anaplasma cf. platys form a separate and well-supported (bootstrap support of 99%) branch on the phylogenetic tree belonging to the cluster of A. platys (Fig. 3). The potentially new species identified from ruminants were also amplified from three samples taken from each ruminant species. The nine sequences from each gene (16S rRNA or rpoB) were identical to each other and showed for the 16S rRNA 97% identity with multiple A. phagocytophilum reported worldwide, multiple A. bovis sequences reported from China and multiple sequences of uncharacterized Anaplasma spp. reported from China and Malaysia. The rpoB sequence of this species showed 79% with A. centrale strain Israel (CP001759), A. phagocytophilum strain Dog 2 (GenBank: CP006618), JM (GenBank: CP006617) and 78% with A. marginale strain Dawn (GenBank: CP006847) and Gypsy Plains (GenBank: CP006846). Sequence analysis of the concatenated 23S rRNA, 16S rRNA and rpoB genes revealed that this Anaplasma species was distinct from other Anaplasmataceae species considering the lower sequence identity (93% for 23S rRNA, 97% for 16S rRNA and 79% for rpoB). Because these potentially new species had not previously been reported, we propose the provisional name “Candidatus Anaplasma africae”. The phylogenetic tree showed that “Candidatus Anaplasma africae” form a separate and well-supported (bootstrap support of 100%) branch on the phylogenetic tree situated between Anaplasma spp. and Ehrlichia spp., albeit closer to the Anaplasma spp. clusters (Fig. 3).
Phylogenetic tree showing the position of A. ovis, A. marginale, Anaplasma cf. platys and “Ca. Anaplasma africae” amplified from ruminates and A. platys amplified from dogs compared to other Anaplasmataceae species. Evolutionary analyses were conducted using MEGA7 [26]. The concatenated 23S rRNA, 16S rRNA and the rpoB genes of the Anaplasma spp. amplified in this study with other sequences of Anaplasmataceae species available from GenBank were aligned using CLUSTAL W implemented on BioEdit v.3 [25] (there were 1599 positions in the final dataset). The evolutionary history was inferred by using the maximum likelihood method based on theHasegawa–Kishino–Yano model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood approach and then selecting the topology with superior log likelihood value. Statistical support for internal branches of the trees was evaluated by bootstrapping with 1000 iterations. A discrete gamma distribution was used to model evolutionary rate differences among sites [2 categories (+G, parameter = 0.3463)]. The analysis involved 41 nucleotide sequences. All positions containing gaps and missing data were excluded. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The scale-bar represents a 20% nucleotide sequence divergence. Accession number for each concatenated sequence in the phylogenetic tree were provided for each species as [23S rRNA, rpoB and 16S rRNA]
The sequences of A. platys 16S rRNA amplified from dogs showed 99% identity with the sequences of A. platys amplified from ruminants in the present study and 99% with multiple sequences of A. platys reported worldwide. However, they also showed 99% identity with “Candidatus Anaplasma camelii” clone Camel_38 (GenBank: KF843827). The rpoB sequence showed 99% identity with A. platys strain Gard 1 (GenBank: KX155493) (Fig. 3). The 16S rRNA sequences of E. canis amplified from dogs and the two R. e. evertsi ticks showed 99% identity with E. canis reported worldwide and 99% identity with “E. ovina” (GenBank: AF318946). The groEL sequences of this E. canis strain showed 99% with E. canis isolate D12E (GenBank: JN391407-JN391408) reported from the Philippines and E. canis strain Jake (GenBank: CP000107) reported from the USA (Fig. 4).
Phylogenetic tree showing the position of E. canis amplified from dogs and Rhipicephalus bursa ticks collected from sheep compared to other Anaplasmataceae species. Evolutionary analyses were conducted using MEGA7 [26]. The concatenated 23S rRNA, groEl and 16S rRNA genes of the E. canis amplified in this study with other sequences of Anaplasmataceae species available from GenBank were aligned using CLUSTAL W implemented on BioEdit v.3 [25] (there were 1818 positions in the final dataset). The evolutionary history was inferred by using the maximum likelihood method based on theHasegawa–Kishino–Yano model. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree for the heuristic search was obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum composite likelihood (MCL) approach and then selecting the topology with superior log likelihood value. Statistical support for internal branches of the trees was evaluated by bootstrapping with 1000 iterations. A discrete gamma distribution was used to model evolutionary rate differences among sites [2 categories (+G, parameter = 0.4640)]. The analysis involved 41 nucleotide sequences. All positions containing gaps and missing data were excluded. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The scale-bar represents a 5% nucleotide sequence divergence
Discussion
The clinical presentation of anaplasmosis depends on multiple factors including the Anaplasmataceae species, strain and host. Other factors associated with immune status and co-infections with other pathogens make the diagnosis a very challenging task. In addition, the economic impact, the zoonotic potential and the presence of multiple vectors associated with the transmission of these bacteria determine the need for accurate and direct laboratory tests [15]. Epidemiological data about the spread of bacteria belonging to the family Anaplasmataceae in addition to the identification of potential reservoir and vectors in the region will facilitate the interpretation of bacteriological results among the infected hosts. The present study summarizes entomological and epidemiological data of the prevalence of Anaplasmataceae species infecting animals and ticks in three regions of Senegal. In Keur Momar Sarr, five different tick species were collected from sheep, goats, cattle, horses, donkeys and dogs. Except for R. bursa, other ticks, namely R. e. evertsi, Hy. rufipes, Hy. impeltatum and R. muhsamae, had already been reported in Senegal and belong to the 33 known species in this country [27]. Walker et al. [20] states that the records of R. bursa from outside the Palaearctic region are linked to misidentification or accidental importation [20]. This species is usually recorded in the Mediterranean area, and some cold regions of Europe including the French Basque country [22] and Crimea [28]. In the present study, three engorged females removed from three horses were identified by molecular characterization as R. bursa. The occurrence of this species was very low 3/204 (1.5%). The data collected in the present study are insufficient to conclude or suggest an accidental introduction or for a possible establishment and/or the presence of foci of these ticks in the arid conditions of the northern region of Senegal.
The most abundant tick species found was R. e. evertsi (84.3%), collected from almost all animal species except from dogs. Rhipicephalus e. evertsi is an Afrotropical species. In West Africa, R. e. evertsi is absent on wild ungulates, suggesting that the species was introduced on domestic livestock from East Africa [20]. This species is the most common tick in southern Senegal [29]. In Africa, R. e. evertsi is present in a band extending roughly from 10–16°N and 11°W to 20°E, a region that receives between 400–1000 mm rainfall annually [20]. Rhipicephalus e. evertsi is known in Senegal as a potential vector of Rickettsia africae, R. aeschlimannii, R. conorii and Coxiella burnetii [27, 29, 30]. In our study, 2/176 (1.1%) R. e. evertsi collected from sheep were infected with E. canis. These two ticks were removed from the same sheep. To the best of our knowledge, this is the first identification of E. canis in R. e. evertsi. In previous studies, A. platys, E. ruminantium and A. ovis were also reported from R. e. evertsi in South Africa and Ethiopia [31,32,33]. However, to our knowledge, E. canis has never been reported from sheep. Rhipicephalus e. evertsi is a two-host tick species with larvae and nymph infesting the first host and adults infesting the second host [18]. Interestingly, R. e. evertsi was shown experimentally to transmit “E. ovina” to sheep by adult ticks. “Ehrlichia ovina” is a not a completely described Ehrlichia species but it is reported to infect domestic ruminants from Turkey and the Caribbean islands [34, 35]. The 16S rRNA sequences of “E. ovina” (GenBank: AF318946) showed 100% identity with multiple E. canis sequences reported worldwide [36], whereas the gltA sequence (GenBank: KP719095) showed 99.9 % (two mismatches) identical to that of E. canis from Italy [34]. Nonetheless, stocks of “E. ovina” are not readily available and their taxonomic position needs to be analyzed for other genes including groEL [1] and rpoB [22]. These phylogenetic analyses are necessary to understand the validity of “E. ovina” as a species or to combine it with E. canis, as well as to confirm and consider R. e. evertsi as a competent vector for E. canis.
Hyalomma rufipes ticks were removed only from cattle and horses. This species represents 8.3% of the overall tick species collected. Hyalomma rufipes is most common in dry areas and distributed in almost all African countries [29]. This tick is found on a large variety of animals include wild mammals and birds and is considered an important vector of the Crimean-Congo hemorrhagic fever [37]. In Senegal, this tick has been identified as a host for R. aeschlimannii and C. burnetii [30]. In South Africa, A. marginale was found in these ticks [32, 38]. None of the Hy. rufipes tested here were infected by Anaplasmataceae species.
Hyalomma impeltatum was collected exclusively from horses at low occurrence (4.9%). In Senegal, Hy. impeltatum is more frequently encountered in cows than in sheep [29]. This Afrotropical tick was also reported infected by R. aeschlimannii [29] and interestingly by Ehrlichia chaffensis in Nigeria [39]. Ehrlichia chaffensis is an emerging bacteria in Africa, and until now, this species has been reported in three countries in Africa including Nigeria, Uganda and Cameroon [39, 40]. Rhipicephalus sanguineus and Hy. impeltatum seem to be potential vectors of E. chaffensis in Africa [39, 41]. In our study, none of the ten Hy. impeltatum collected were infected by an Anaplasmataceae species. However, E. chaffensis has never been reported from humans or other animals in Senegal. Finally, only two R. muhsamae (0.9%) were collected exclusively from dogs. Rhipicephalus muhsamae is an Afrotropical species found mainly in forest savanna mosaic and less abundant in tropical and subtropical moist broadleaf forest [18]. Rickettsia conorii has been detected in R. mushamae ticks removed from cattle in the Central African Republic [42]. None of the two R. mushamae collected were infected by Anaplasmataceae species.
Four hundred thirty-three blood samples were collected from different animals including sheep, goats, cattle, horses and dogs. The overall prevalence of Anaplasmataceae infection in the sampled animals was 41.1%. Anaplasma ovis was the common species found in sheep (55.9%). The infection prevalence found in the present study was higher compared to that previously reported in Senegal in 2013 (31.6%) [43]. Sheep develop persistent infections which allow them to become reservoirs of infection, explaining the high rate of infection by A. ovis in sheep [5]. The infection is usually subclinical and associated with hemolytic anemia. Cross-infection with other parasites or other stress conditions increase the severity of the infection [14]. Complication with other opportunistic diseases or stress conditions in sheep infected by A. ovis lead to the development of an acute disease phase characterized by fever, progressive anemia, icterus, weight loss, milk yield decrease and sometimes death [44]. In Africa, A. ovis has been reported from sheep in Ethiopia, South Africa, Tunisia [33, 44, 45] and from goats in Angola [46]. Rhipicephalus e. evertsi was previously reported as an important vector of A. ovis in Africa [37, 38]. However, none of the R. e. evertsi collected here was infected by A. ovis. Anaplasma marginale and A. centrale were identified in 19.4 and 8.1% of cattle, respectively. These two pathogens have never been reported in Senegal but have been reported from many other countries in Africa including Algeria, Angola, Botswana, Egypt, Ivory Coast, Kenya, Morocco, Mozambique, Nigeria, South Africa, Sudan, Tanzania, Tunisia, Uganda and Zambia [47,48,49,50,51,52,53,54]. The vectors of these two pathogens in Africa were ticks belonging to the genera Rhipicephalus and Amblyomma [5, 52]. The prevalence of these bacteria in cattle reported from the eastern and southern countries of Africa ranges between 32.1–100% [51]. Infection with Anaplasma marginale is associated with mild to severe anemia. Cattle infected with this bacterium developed various clinical signs including fever, decline in milk production, temporary infertility, and some animals developed an acute disease that manifested with gastrointestinal and neurological signs associated with the development of icterus seen during early convalescence. Mortality rates can reach 50–60% in adult cattle [5, 12]. Anaplasma centrale is less pathogenic and has been used as a live blood vaccine to protect against bovine anaplasmosis caused by A. marginale [55].
Anaplasma cf. platys found in this study was gentically close to the dog pathogen A. platys. Many genotypes close to A. platys, commonly named Anaplasma platys-like bacteria, were reported from animal families other than dogs [56]. Anaplasma platys-like is considered an emerging bacterium in many countries of the world. The same bacterium was reported previously from two sheep in Senegal [43]. In the present study, the prevalence found was 27.7% in goats, 22.6% in cattle and 19.8% in sheep. This species was initially characterized from ruminants in Italy [57]. Then, Anaplasma cf. platys (the commonly called Anaplasma platys-like) was reported from cattle in Algeria and Tunisia, from sheep in South Africa and goats in China [23, 45, 58, 59]. Recently this infection was also characterized from cats in Italy [56]. This species is described as a neutrophil-tropic Anaplasma sp. in ruminants and platelet-tropic in cats [57]. The strain identified here is genetically closest to the canine A. platys (nucleotide homology was 99% for rrs and rrl); however, the rpoB gene showed 93% identity with A. platys. Anaplasma cf. platys is located independently in a separate group in the phylogenetic tree based on the concatenated genes rrl, rrs and rpoB (Fig. 2).
The putative new species identified here, provisionally named “Ca. Anaplasma africae”, has genetic features which are different from all other species of the genus Anaplasma. A phylogenetic tree based on the concatenated rrl, rrs and rpoB genes showed that “Ca. Anaplasma africae” forms a separate branch on the phylogenetic tree situated between Anaplasma and Ehrlichia, albeit closer to the Anaplasma clusters (Fig. 2). The prevalence of this species is low in goats, cattle and sheep, 10.3%, 8.1% and 3.7%, respectively. The importance of Anaplasma cf. platys and “Ca. Anaplasma africae” in ruminants has to be elucidated.
Ehrlichia canis and A. platys were identified exclusively from dogs sampled in the Keur Momar Sarr villages. None of the dogs sampled in the Casamance region were found positive. In dogs from Keur Momar Sarr, the prevalence observed was 18.8% for E. canis and 15.6% for A. platys. The prevalence of E. canis reported in the present study was low compared to previous reports in dogs sampled in a kennel in Dakar, Senegal (53%). However, the prevalence of A. platys was higher than what was reported in the same study (5.9%) [60]. Ehrlichia canis infection in dogs induces monocytotropic ehrlichiosis and results in various signs depending on the disease stages [61], whereas A. platys induces a recurrent thrombocytopenia that can be resolved in the absence of other infecting agents or complicating factors [62, 63]. The prevalence to E. canis and A. platys in Africa is poorly known; however, these two species are apparently ubiquitous throughout the African area where the R. sanguineus tick group is spread [17, 64].
Nonetheless, dogs from the Central African Republic and Reunion Island were all negative for E. canis [65]. Rhipicephalus sanguineus (s.l.) in the tropical and subtropical region are active throughout the year and apparently have no seasonality [66]. In our study, two R. mushamae ticks were collected from two dogs. To the best of our knowledge, A. platys has never been associated with R. mushamae. Ehrlichia canis has been reported once from R. mushamae in Mali [17]. In Africa, from tick species other than R. sanguineus (s.l.), A. platys was amplified from R. e. evertsi salivary glands in South Africa [32] and R. camicasi in Kenya and Ivory Coast [66].
None of the 64 horses tested were found positive for an Anaplasmataceae infection. In addition, none of the Anaplasmataceae species identified from ruminants and dogs were identified in ticks, except for E. canis identified in dogs and R. e. evertsi collected from sheep. Anaplasma cf. platys and “Ca. Anaplasma africae” were found in sheep, goats and cattle. Despite the fact that R. e. evertsi were collected from different ruminant animals, none of these ticks were found positive for any Anaplasmataceae species. Anaplasma phagocytophilum was also not found in the present study. Anaplasma phagocytophilum has already been identified in sheep in Senegal, although all of its known vectors are absent in sub-Saharan Africa [43]. Evidence of the presence of A. phagocytophilum in Africa has been reported from Morocco, Algeria and Tunisia [23, 54, 67], where Ixodes ricinus has been identified in some foci in the northern regions of these countries [18]. This infection has also been reported from wildlife in Zimbabwe and South Africa [68, 69], and from some African ticks such as Ambyomma flavomaculatum collected from two different specimens of lizard (Varanus exanthematicus) imported to Poland from Ghana [70], from A. cohaerens, A. lepidum and A. variegatum in Ethiopia [71, 72] and from Hy. marginatum in Tunisia [53]. In our study, none of the five tick species were found to be infected by A. phagocytophilum, suggesting the implication of other tick species in the epidemiology of A. phagocytophilum in sub-Saharan Africa.
Conclusions
The present study aimed to identify Anaplasmataceae species infecting animals and ticks and determine a possible epidemiological Anaplasmataceae-host-tick connection. The present work indicates that ruminants and dogs in the northern and central areas of Senegal are a reservoir for multiple Anaplasmataceae species. The prevalence of A. ovis and A. marginale was high in sheep and cattle, respectively. Molecular analysis revealed an interesting diversity of Anaplasmataceae infections in ruminants and dogs including a potentially new species infecting ruminants. However, except for E. canis, none of the other Anaplasma spp. found in the present study was amplified from ticks. Nevertheless, further studies are needed to ascertain the Anaplasmataceae-host-vector connections in sub-Saharan Africa as well as to decipher the zoonotic potential of newly identified genotypes and their significance for animals and public health.
Availability of data and materials
All data are available from the corresponding author upon reasonable request. Representative sequences were submitted to the GenBank database under the following accession numbers. The 23S rRNA gene sequences: A. platys DNS46 [MN317236], A. ovis SNS2 [MN317237], A. cf. platys from sheep (SNS84), goats (GNS15) and cattle (CNS12) [MN317238, MN317239 and MN317240, respectively], “Ca. Anaplasma africae” from sheep (SNS93), goats (GNS26) and cattle (CNS7) [MN317241, MN317242 and MN317243, respectively], A. marginale CNS5 [MN317244], A. centrale CNS11 [MN317245], E. canis from R. bursa [MN317246] and dogs [MN317247]. The rpoB gene sequences: A. platys DNS46 [MN284923], A. ovis SNS2 [MN284924], A. cf. platys from sheep (SNS84), goats (GNS15) and cattle (CNS12) [MN284925, MN284926 and MN284927, respectively], “Ca. Anaplasma africae” from sheep (SNS93), goats (GNS26) and cattle (CNS7) [MN284928, MN284929 and MN284930, respectively], A. marginale CNS5 [MN284931], A. centrale CNS1 [MN284932]. The GroEL gene sequences: E. canis from dogs [MN216188], E. canis from R. bursa [MN216187]. The 16S rRNA gene sequences: A. platys DNS46 [MN317248], A. ovis SNS2 [MN317249], A. cf. platys from sheep (SNS84), goats (GNS15) and cattle (CNS12) [MN317250, MN317251 and MN317252, respectively], “Ca. Anaplasma africae” from sheep (SNS93), goats (GNS26) and cattle (CNS7) [MN317253, MN317254 and MN317255, respectively], A. marginale CNS5 [MN317256], A. centrale CNS11 [MN317257], E. canis from R. bursa [MN317258] and dogs [MN317259]. The tick 12S rRNA gene sequences: R. evertsi evertsi from sheep, equines, donkeys and cattle [MN315378, MN315379, MN315380 and MN315381, respectively]; H. rufipes from equines and cattle [MN315382 and MN315383, respectively]; H. impeltatum from equines [MN315384]; R. bursa from equines [MN315385]; and R. muhsamae from dogs [MN315386].
Abbreviations
- Ap-ha:
-
Anaplasma phagocytophilum strain human-active
- Ap-v1:
-
Anaplasma phagocytophilum variant 1
- SNP:
-
single-nucleotide polymorphism
References
Dumler JS, Barbet AF, Bekker CP, Dasch G, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‛HGE agentʼ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–65.
Greiman SE, Rikihisa Y, Cain J, Vaughan JA, Tkach VV. Germs within worms: localization of Neorickettsia sp. within life cycle stages of the digenean Plagiorchis elegans. Appl Environ Microbiol. 2016;82:2356–62.
de la Fuente J, Kocan KM, Blouin EF, Zivkovic Z, Naranjo V, Almazán C, et al. Functional genomics and evolution of tick-Anaplasma interactions and vaccine development. Vet Parasitol. 2010;167:175–86.
Rar V, Golovljova I. Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect Genet Evol. 2011;11:1842–61.
Kocan KM, de la Fuente J, Blouin EF, Coetzee JF, Ewing SA. The natural history of Anaplasma marginale. Vet Parasitol. 2010;167:95–107.
de la Fuente J, Estrada-Peña A, Cabezas-Cruz A, Kocan KM. Anaplasma phagocytophilum uses common strategies for infection of ticks and vertebrate hosts. Trends Microbiol. 2016;24:173–80.
Rar VA, Epikhina TI, Livanova NN, Panov VV, Doroshenko EK, Pukhovskaia NM, et al. Study of the heterogeneity of 16S rRNA gene and groESL operone in the DNA samples of Anaplasma phagocytophilum, Ehrlichia muris, and “Candidatus Neoehrlichia mikurensis” determined in the Ixodes persulcatus ticks in the area of Urals, Siberia, and Far East of Russia. Mol Gen Mikrobiol Virusol. 2011;26:17–23.
Dugat T, Lagrée A, Maillard R, Boulouis H-J, Haddad N. Opening the black box of Anaplasma phagocytophilum diversity: current situation and future perspectives. Front Cell Infect Microbiol. 2015;5:61.
Keesing F, Mchenry DJ, Hersh M, Tibbetts M, Brunner JL, Killilea M, et al. Prevalence of human-active and variant 1 strains of the tick-borne pathogen Anaplasma phagocytophilum in hosts and forests of eastern North America. Am J Trop Med Hyg. 2014;91:302–9.
Durrheim DN, Freeman P, Roth I, Hornitzky M. Distinct ecologically relevant strains of Anaplasma phagocytophilum. Emerg Infect Dis. 2009;15:842–3.
Stuen S, Granquist EG, Silaghi C. Anaplasma phagocytophilum - a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect Microbiol. 2013;3:31.
Aubry P, Geale DW. A review of bovine anaplasmosis. Transbound Emerg Dis. 2011;58:1–30.
Nair AS, Ravindran R, Lakshmanan B, Sreekumar C, Kumar SS, Raju R, et al. Bovine carriers of Anaplasma marginale and Anaplasma bovis in South India. Trop Biomed. 2013;30:105–12.
Alessandra T, Santo C. Tick-borne diseases in sheep and goats: clinical and diagnostic aspects. Small Rumin Res. 2012;106:S6–11.
Silaghi C, Santos AS, Gomes J, Christova I, Matei IA, Walder G, et al. Guidelines for the direct detection of Anaplasma spp. in diagnosis and epidemiological studies. Vector-Borne Zoonotic Dis. 2017;17:12–22.
Li H, Zheng Y-C, Ma L, Jia N, Jiang B-G, Jiang R-R, et al. Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect Dis. 2015;15(6):663–7.
Ehounoud CB, Fenollar F, Dahmani M, N’Guessan JD, Raoult D, Mediannikov O. Bacterial arthropod-borne diseases in West Africa. Acta Trop. 2017;7:124–37.
Guglielmone AA, Robbins RG, Apanaskevich DA, Petney TN, Estrada-Peña A, Horak IG. The hard ticks of the world (Acari: Ixodida: Ixodidae). Dordrecht: Springer; 2014.
Estrada-Peña A, Bouattour A, Camicas JL, Walker AR. Ticks of domestic animals in the Mediterranean region. Zaragoza: University of Zaragoza; 2004.
Walker A. The genus Rhipicephalus (Acari, Ixodidae): A guide to the brown ticks of the World. In: Walker JB, Keirans JE, Horak IG, editors. Tropical animal health production. Dordrecht: Kluwer Academic Publishers; 2000. p. 417–8.
Beati L, Keirans JE. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J Parasitol. 2001;87:32–48.
Dahmani M, Davoust B, Rousseau F, Raoult D, Fenollar F, Mediannikov O. Natural Anaplasmataceae infection in Rhipicephalus bursa ticks collected from sheep in the French Basque Country. Ticks Tick Borne Dis. 2017;8:18–24.
Dahmani M, Davoust B, Benterki MS, Fenollar F, Raoult D, Mediannikov O. Development of a new PCR-based assay to detect Anaplasmataceae and the first report of Anaplasma phagocytophilum and Anaplasma platys in cattle from Algeria. Comp Immunol Microbiol Infect Dis. 2015;4:39–45.
Dahmani M, Loudahi A, Mediannikov O, Fenollar F, Raoult D, Davoust B. Molecular detection of Anaplasma platys and Ehrlichia canis in dogs from Kabylie, Algeria. Ticks Tick Borne Dis. 2015;6:198–203.
Hall TA. BioEdit: a user-friendly biological sequences alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series No. 41. Oxford: Oxford University Press; 1999. p. 95–8.
Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 70 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Mediannikov O, Diatta G, Fenollar F, Sokhna C, Trape JF, Raoult D. Tick-borne rickettsioses, neglected emerging diseases in rural Senegal. PLoS Negl Trop Dis. 2010;4:e821.
Akimov IA, Nebogatkin IV. Ticks of the genus Rhipicephalus (Acari, Ixodidae) and their distribution in Ukraine. Vestn Zool. 2013;47:28–34.
Sambou M, Faye N, Bassène H, Diatta G, Raoult D, Mediannikov O. Identification of rickettsial pathogens in ixodid ticks in northern Senegal. Ticks Tick Borne Dis. 2014;5:552–6.
Mediannikov O, Fenollar F, Socolovschi C, Diatta G, Bassene H, Molez JF, et al. Coxiella burnetii in humans and ticks in rural Senegal. PLoS Negl Trop Dis. 2010;4:e654.
Mtshali K, Khumalo ZTH, Nakao R, Grab DJ, Sugimoto C, Thekisoe OMM. Molecular detection of zoonotic tick-borne pathogens from ticks collected from ruminants in four South African provinces. J Vet Med Sci. 2015;77:1573–9.
Berggoetz M, Schmid M, Ston D, Wyss V, Chevillon C, Pretorius A, et al. Protozoan and bacterial pathogens in tick salivary glands in wild and domestic animal environments in South Africa. Ticks Tick Borne Dis. 2014;5:176–85.
Teshale S, Kumsa B, Menandro ML, Cassini R, Martini M. Anaplasma, Ehrlichia and rickettsial pathogens in ixodid ticks infesting cattle and sheep in western Oromia, Ethiopia. Exp Appl Acarol. 2016;70:231–7.
Zhang J, Kelly P, Guo W, Xu C, Wei L, Jongejan F, et al. Development of a generic Ehrlichia FRET-qPCR and investigation of ehrlichioses in domestic ruminants on five Caribbean islands. Parasit Vectors. 2015;8:506.
Kelly PJ, Matthewman LA, Mahan SM, Semu S, Peter T, Mason PR, et al. Serological evidence for antigenic relationships between Ehrlichia canis and Cowdria ruminantium. Res Vet Sci. 1994;56:170–4.
Bekker CPJ, De Vos S, Taoufik A, Sparagano OAE, Jongejan F. Simultaneous detection of Anaplasma and Ehrlichia species in ruminants and detection of Ehrlichia ruminantium in Amblyomma variegatum ticks by reverse line blot hybridization. Vet Microbiol. 2002;89:223–38.
Chen Z, Li Y, Liu Z, Yang J, Yin H. The life cycle of Hyalomma rufipes (Acari: Ixodidae) under laboratory conditions. Exp Appl Acarol. 2012;56:85–92.
Mutshembele AM, Cabezas-Cruz A, Mtshali MS, Thekisoe OMM, Galindo RC, de la Fuente J. Epidemiology and evolution of the genetic variability of Anaplasma marginale in South Africa. Ticks Tick Borne Dis. 2014;5:624–31.
Reye AL, Arinola OG, Hübschen JM, Muller CP. Pathogen prevalence in ticks collected from the vegetation and livestock in Nigeria. Appl Environ Microbiol. 2012;78:2562–8.
Proboste T, Kalema-Zikusoka G, Altet L, Solano-Gallego L, Mera IG, Chirife AD, et al. Infection and exposure to vector-borne pathogens in rural dogs and their ticks, Uganda. Parasit Vectors. 2015;8:306.
Ndip LM, Ndip RN, Esemu SN, Walker DH, McBride JW. Predominance of Ehrlichia chaffeensis in Rhipicephalus sanguineus ticks from kennel-confined dogs in Limbe, Cameroon. Exp Appl Acarol. 2010;50:163–8.
Parola P, Paddock CD, Labruna MB, Mediannikov O, Abdad MY, Stenos J, et al. Update on tick-borne rickettsioses around the world : a geographic approach. Clin Microbiol Rev. 2013;26:657–702.
Djiba ML, Mediannikov O, Mbengue M, Thiongane Y, Molez J-F, Seck MT, et al. Survey of Anaplasmataceae bacteria in sheep from Senegal. Trop Anim Health Prod. 2013;45:1557–61.
Belkahia H, Said M, Alberti A, Abdi K, Issaoui Z, Hattab D, et al. First molecular survey and novel genetic variants’ identification of Anaplasma marginale, A centrale and A bovis in cattle from Tunisia. Infect Genet Evol. 2015;34:361–71.
Berggoetz M, Schmid M, Ston D, Wyss V, Chevillon C, Pretorius AM, Gern L. Tick-borne pathogens in the blood of wild and domestic ungulates in South Africa: interplay of game and livestock. Ticks Tick Borne Dis. 2014;5:166–75.
Kubelova M, Mazancova J, Siroky P. Theileria, Babesia, and Anaplasma detected by PCR in ruminant herds at Bie Province, Angola. Parasite. 2012;19:417–22.
El-ashker M, Hotzel H, Gwida M, El-beskawy M, Silaghi C, Tomaso H. Molecular biological identification of Babesia, Theileria, and Anaplasma species in cattle in Egypt using PCR assays, gene sequence analysis and a novel DNA microarray. Vet Parasitol. 2015;207:329–34.
Ait Hamou S, Rahali T, Sahibi H, Belghyti D, Losson B, Goff W, et al. Molecular and serological prevalence of Anaplasma marginale in cattle of North Central Morocco. Res Vet Sci. 2012;93:1318–23.
Eygelaar D, Jori F, Mokopasetso M, Sibeko KP, Collins NE, Vorster I, et al. Tick-borne haemoparasites in African buffalo (Syncerus caffer) from two wildlife areas in northern Botswana. Parasit Vectors. 2015;8:26.
Lorusso V, Wijnveld M, Majekodunmi AO, Dongkum C, Fajinmi A, Dogo AG, et al. Tick-borne pathogens of zoonotic and veterinary importance in Nigerian cattle. Parasit Vectors. 2016;9:217.
Tembue AAM, Da Silva JB, Da Silva FJM, Pires MS, Baldani CD, Soares CO, et al. Seroprevalence of IgG antibodies against Anaplasma marginale in cattle from south Mozambique. Rev Bras Parasitol Veterinária. 2011;20:318–24.
Ehounoud C, Yao K, Dahmani M, Achi Y, Amanzougaghene N, N’Douba AK, et al. Multiple pathogens including potential new species in tick vectors in Côte d’Ivoire. PLoS Negl Trop Dis. 2016;10:e0004367.
M’Ghirbi Y, Yaïch H, Ghorbel A, Bouattour A. Anaplasma phagocytophilum in horses and ticks in Tunisia. Parasit Vectors. 2012;5:180.
M’Ghirbi Y, Bèji M, Oporto B, Khrouf F, Hurtado A, Bouattour A. Anaplasma marginale and A. phagocytophilum in cattle in Tunisia. Parasit Vectors. 2016;9:556.
Bell-Sakyi L, Palomar AM, Bradford EL, Shkap V. Propagation of the Israeli vaccine strain of Anaplasma centrale in tick cell lines. Vet Microbiol. 2015;179:270–6.
Zobba R, Anfossi AG, Visco S, Sotgiu F, Dedola C, Parpaglia MLP, et al. Cell tropism and molecular epidemiology of Anaplasma platys-like strains in cats. Ticks Tick Borne Dis. 2015;6:272–80.
Zobba R, Anfossi AG, Pinna Parpaglia ML, Dore GM, Chessa B, Spezzigu A, et al. Molecular investigation and phylogeny of Anaplasma spp in Mediterranean ruminants reveal the presence of neutrophil-tropic strains closely related to A. platys. Appl Environ Microbiol. 2014;80:271–80.
Ben Said M, Belkahia H, El N, Saidani M, Alberti A, Zobba R, et al. Anaplasma platys-like strains in ruminants from Tunisia. Infect Genet Evol. 2017;49:226–33.
Zhang L, Liu H, Xu B, Lu Q, Li L, Chang L, et al. Anaplasma phagocytophilum infection in domestic animals in ten provinces/cities of China. Am J Trop Med Hyg. 2012;87:185–9.
Davoust B, Mediannikov O, Chene J, Massot R, Tine R, Diarra M, et al. Study of ehrlichiosis in kennel dogs under treatment and prevention during seven months in Dakar (Senegal). Comp Immunol Microbiol Infect Dis. 2013;36:613–7.
Harrus S, Waner T. Diagnosis of canine monocytotropic ehrlichiosis (Ehrlichia canis): an overview. Vet J. 2011;187:292–6.
Gaunt SD, Beall MJ, Stillman BA, Lorentzen L, Diniz PPVP, Chandrashekar R, Breitschwerdt EB. Experimental infection and co-infection of dogs with Anaplasma platys and Ehrlichia canis: hematologic, serologic and molecular findings. Parasit Vectors. 2010;3:33.
Diniz PPVP, Beall MJ, Omark K, Chandrashekar R, Daniluk DA, Cyr KE, et al. High prevalence of tick-borne pathogens in dogs from an Indian reservation in northeastern Arizona. Vector Borne Zoonotic Dis. 2010;10:117–23.
Davoust B, Parzy D, Demoncheaux J-P, Tine R, Diarra M, Marié J-L, et al. Usefulness of a rapid immuno-migration test for the detection of canine monocytic ehrlichiosis in Africa. Comp Immunol Microbiol Infect Dis. 2014;37:31–7.
Socolovschi C, Gomez J, Marié J, Davoust B, Guigal PM, Raoult D, et al. Ehrlichia canis in Rhipicephalus sanguineus ticks in the Ivory Coast. Ticks Tick Borne Dis. 2012;3:411–3.
Matei IA, D’Amico G, Yao PK, Ionică AM, Kanyari PWN, Daskalaki AA, et al. Molecular detection of Anaplasma platys infection in free-roaming dogs and ticks from Kenya and Ivory Coast. Parasit Vectors. 2016;9:157.
Elhamiani Khatat S, Sahibi H, Hing M, Alaoui Moustain I, El Amri H, Benajiba M, et al. Human exposure to Anaplasma phagocytophilum in two cities of northwestern Morocco. PLoS ONE. 2016;11:e0160880.
Nakayima J, Hayashida K, Nakao R, Ishii A, Ogawa H, Nakamura I, et al. Detection and characterization of zoonotic pathogens of free-ranging non-human primates from Zambia. Parasit Vectors. 2014;7:490.
Kelly P, Marabini L, Dutlow K, Zhang J, Loftis A, Wang C. Molecular detection of tick-borne pathogens in captive wild felids, Zimbabwe. Parasit Vectors. 2014;7:514.
Nowak M, Cieniuch S, Stan J. Detection of Anaplasma phagocytophilum in Amblyomma flavomaculatum ticks (Acari : Ixodidae) collected from lizard Varanus exanthematicus imported to Poland. Exp Appl Acarol. 2010;51:363–71.
Hornok S, Sándor H, Bichu GA, Akács NT, Yuranecz MG. Molecular screening for Anaplasmataceae in ticks and tsetse flies from Ethiopia. Acta Vet Hung. 2016;64:65–70.
Teshale S, Geysen D, Ameni G, Asfaw Y, Berkvens D. Improved molecular detection of Ehrlichia and Anaplasma species applied to Amblyomma ticks collected from cattle and sheep in Ethiopia. Ticks Tick Borne Dis. 2015;6:1–7.
Dahmani M, Marié J-L, Scandola P, Brah S, Davoust B, Mediannikov O. Anaplasma ovis infects sheep in Niger. Small Rumin Res. 2017;151:32–5.
Acknowledgements
We thank Paul Sambou for their assistance during the fieldwork. We also thank Mackenzie Tardif for his proofreading of the manuscript.
Funding
This study was supported by the Institut Hospitalo-Universitaire (IHU) Méditerranée Infection, the National Research Agency under the program “Investissements d’avenir”, reference ANR-10-IAHU-03, the Région Provence Alpes Côte d’Azur and European funding FEDER PRIMI. Funding sources had no role in the design or conduct of the study; collection, management, analysis or interpretation of the data; or preparation, review or approval of the manuscript.
Author information
Authors and Affiliations
Contributions
OM, BD, MD, FF and DR designed the study. BD, MS, HB, PS and OM collected samples in Senegal. MD and TA performed the laboratory analyses. MD and OM carried out the data analysis. MD and OM drafted the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Animal blood samples were collected according to good practice by veterinarians and according to Senegalese regulations with the agreement of the owners.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Dahmani, M., Davoust, B., Sambou, M. et al. Molecular investigation and phylogeny of species of the Anaplasmataceae infecting animals and ticks in Senegal. Parasites Vectors 12, 495 (2019). https://doi.org/10.1186/s13071-019-3742-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-019-3742-y