Abstract
Background
Four species within Anaplasma genus are emerging zoonotic pathogens, which are transmitted by ticks and generate veterinary and public health concerns. Here, we performed a molecular survey of Anaplasma in Ankang, Northwest China.
Methods
Hard ticks were collected and identified using morphological and molecular methods. Human-pathogenic Anaplasma species were tested using nested polymerase chain reaction. The nearly complete rrs, gltA, and groEL genes sequences from revealed Anaplasma species were amplified and sequenced to determine their molecular characteristics and their phylogeny.
Results
All ticks collected in Ankang belonged to the Rhipicephalus microplus. Novel unclassified Anaplasma strains genetically related to A. platys and A. capra were detected in these ticks. Co-infection of these two organisms was also found. The novel unclassified Anaplasma strains identified in this study formed a distinct phylogenetic lineage based on the groEL gene and two lineages based on the gltA gene within A. platys and related strains group. The revealed A. capra strains identified in this study were most closely related to those detected in humans and other vertebrate animals.
Conclusion
We revealed the presence of A. capra, a novel human pathogens in R. microplus ticks in previously unrecognized endemic regions. We also detected a novel unclassified Anaplasma species genetically related to A. platys. The epidemiology of anaplasmosis caused by these two Anaplasma species in humans should be assessed in future studies.
Similar content being viewed by others
Background
Anaplasma genus comprises a group of Gram-negative obligate intracellular bacteria that inhabit diverse eukaryotic hosts. Seven classified species have been described in this genus so far [1,2,3]. In the last two decades, the rapidly spreading diseases caused by Anaplasma in humans and livestock have received increasing attention since the first incidence of human granulocytic anaplasmosis (HGA) caused by A. phagocytophilum was reported [4]. During this period, many known and emerging Anaplasma species have been proved to be pathogenic to humans, causing anaplasmosis or associated diseases. For instance, A. ovis and A. platys have been identified as human pathogens [5, 6], and A. capra, a novel Anaplasma species, was also confirmed to be a human pathogen in China [3]. Among these causative agents, A. phagocytophilum is the most common pathogen with a worldwide distribution [1, 7], while others are usually neglected because the cases occur only sporadically.
Since 1982, several classified species and unclassified strains belonging to Anaplasma genus have been reported in ticks and mammals from China [3, 8, 9]. Human-pathogenic species and unclassified strains within Anaplasma have been identified, including A. capra, A. ovis, A. phagocytophlum, and A. platys. To date, only HGA caused by A. phagocytophilum has been reported [10]. In contrast, less attention has been paid to the epidemiology and genetic characteristics of other pathogens, although they have been reported in ticks and livestock. In Northwest China, molecular epidemiological surveys confirmed the presence of the four Anaplasma species mentioned above: A. phagocytophilum and A. ovis in Qinghai Province; A. ovis in Tibet [11]; A. platys, A. ovis, and A. capra in Gansu Province [12,13,14]; and A. platys and A. ovis in Xinjiang Uygur Autonomous Region [15,16,17].
Shaanxi Province is located in Northwest China, forming a gateway to four other provinces in this region and connecting the East, Central, and Northwest China. Previous studies have revealed the presence of at least 26 tick species in this region [18], but none of them has attempted to molecularly characterize the human-pathogenic Anaplasma species that they may host. To know more about the circulation and the genetic characteristic of human-pathogenic Anaplasma species in Shaanxi, we sampled ticks from domestic animals in Ankang city and screened them for Anaplasma spp.
Methods
Collection of tick samples
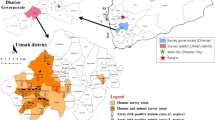
Ankang city is located in Northwest China, southeast of Shaanxi Province and south of the Qinling Mountains (31°42′ to 33°50′N, 108°00′ to 110°12′E). Its topography is characterized by densely vegetated mountains and hills, and livestock often become heavily infested with ticks when they graze in the field. In this study, ticks were sampled in 2017 and 2018 from the body surface of cattle, goats, or sheep, and one tick per animal was collected from seven sampling sites (Fig. 1). These animal hosts of ticks were privatedly-owned and verbal consent was obtained from the owners of these animals for tick samples to be taken. The ticks were first identified by morphology [19, 20], and the identification of one-quarter of adults and all nymphs was further confirmed by analyzing 710-bp of the COI gene using the LCO1490 and HCO2198 primers [21]. The Institutional Animal Care and Use Committee of Northwest A&F University have reviewed and approved this study.
Map with the coordinates of all seven sampling sites (★) in Ankang, Shaanxi Province, China. The maps of Ankang city and Shaanxi Province was generated using ArcGIS (version 10.0; http://www.arcgis.com/index.html) and edited using Adobe Illustrator. The map of China was obtained from http://english.freemap.jp/item/asia/china.html
DNA extraction and detection of Anaplasma species
The total DNA was extracted from the whole body of each tick. The ticks were first washed with 75% alcohol and then with phosphate-buffered saline. After washing, the total DNA was extracted from individual ticks using Tissue DNA Kit (Omega, Norcross, GA, USA) according to the manufacturer’s instructions. The DNA samples were diluted in 80 μL of the elution buffer and stored at − 20 °C.
The extracted DNA samples were initially screened for the presence of Anaplasma spp. by amplifying the rrs gene using nested polymerase chain reaction (PCR) with primers specific for A. phagocytophilum, A. ovis, and A. capra as described previously [22,23,24,25,26]. The primers PLATYS [27], Ehr2, and Ehr4 [28] were used to detect A. platys by semi-nested PCR, also targeting the rrs gene. The primer sequences are shown in the Table 1. Double-distilled water was used as a negative control, and the extracted DNA samples positive for these four Anaplasma species were used as positive controls. To avoid contamination, the DNA extraction, PCR mixture preparation, template addition, and agarose gel electrophoresis were performed in a fume hood in four separate rooms and filter tips were used in these processes.
Amplification of nearly complete rrs, and partial/nearly complete groEL and gltA genes
The positive samples were used to amplify the nearly complete rrs gene, and the molecular characteristics of the revealed Anaplasma strains were analyzed using partial and/or nearly complete groEL and gltA genes. The nearly complete rrs, gltA, and groEL genes were obtained by amplifying two overlapping fragments by semi-nested PCR (Additional file 1: Figure S1).
For A. capra, the former fragment was amplified with fD1/Ehr2 primer pairs for the primary round and fD1/Ehr4 for the secondary round of semi-nested PCR, and the latter fragment was amplified with Ehr1/rp2 for the primary round and Ehr3/rp2 for the secondary round of semi-nested PCR [25, 28]. The same strategy was used to amplify the almost full-length rrs gene of A. platys and related strains, and only the primer PLATYS primer was used instead of fD1.
The partial gltA gene of A. platys and related strains was amplified with Pglt-F/Pglt-R1 primers for the primary round and Pglt-F/Pglt-R2 for the secondary round of semi-nested PCR. The partial groEL gene were amplified with Pgro-F1/Pgro-R primers for the primary round and Pgro-F2/Pgro-R for the secondary round of semi-nested PCR (Additional file 1: Figure S1).
The nearly complete groEL and gltA genes of A. capra were obtained according to Guo et al. [29]. By amplifying two overlapping fragments, almost-full-length groEL and gltA genes of A. platys and related strains were amplified using semi-nested PCR (Additional file 1: Figure S1). The former fragment of gltA was amplified with Pglt-F/Pglt-R1 primer pairs for the primary round and Pglt-F/Pglt-R2 for the secondary round of semi-nested PCR, whereas the latter fragment of gltA was amplified with Pglt-L-F1/Pglt-L-R for the primary round and Pglt-L-F2/Pglt-L-R for the secondary round. For the nearly complete groEL gene, the former fragment was amplified with Pgro-F-F/Pgro-F-R1 for the primary round and Pgro-F-F/Pgro-F-R2 for the secondary round of semi-nested PCR, and the latter fragment was amplified with Pgro-L-F1/Pgro-L-R for the primary round and Pgro-L-F2/Pgro-L-R for the secondary round of semi-nested PCR. All novel primer sequences in this study (Table 2) were designed based on the known sequences of each Anaplasma species.
Cloning and sequencing of the PCR products
The amplified DNA was electrophoresed in 1.0% agarose gels. All the PCR amplicons of expected size were purified using Gel Extraction kit (TaKaRa, Dalian, China). Purified PCR products were ligated with the cloning vector pMD19-T (TaKaRa, Dalian, China), and the recombinant vector was transformed into competent E. coli cells. Finally, positive clones were identified by PCR and at least three positive clones were sequenced for each PCR product to determine the consensus sequence.
Analysis of obtained sequences
Two overlapping fragments were edited and assembled using Bioedit v.7.1.11 to obtain the nearly complete gene sequences [30]. Multiple alignments of DNA sequences of the recovered rrs, gltA, and groEL genes were performed using the ClustalW method in the MEGA 7.0 software with reference sequences from GenBank [31]. The nucleotide identities were calculated using the MegAlign program in Lasergene [32]. The best-fit nucleotide substitution model was determined using MEGA 7.0 to reconstruct the phylogenetic trees based on rrs, gltA and groEL [31]. Phylogenetic trees based on the maximum likelihood method were reconstructed by PhyML v3.2 [33], with 100 replicates for bootstrap analysis. The trees were midpoint-rooted and visualized by MEGA 7.0, presenting a bootstrap value of more than 70%. The sequences that were newly generated for phylogenetic analysis in this study were submitted to GenBank and assigned accession numbers MH716407–MH716436 and MH762071–MH762085.
Results
Collection of samples and identification of tick species
In the summer and autumn of 2017 and 2018, 397 adult and 59 nymph ticks were collected from livestock (65 sheep, 113 goats, and 278 cattle) in rural villages of Ankang, China. Based on the morphology, all collected ticks were identified as Rhipicephalus microplus, and the identification was confirmed molecularly by amplifying the COI gene isolated from 59 nymph and 100 adult ticks. After sequencing of the PCR products, these sequences shared the highest identities (99.3–99.6%) with those of R. microplus, confirming that all collected ticks belonged to this species. Seven representative COI gene sequences selected from each group sharing 100% identities were submitted to GenBank as MK371424–MK371430.
Identification and genetic analyses of Anaplasma spp. in ticks
In the screening of Anaplasma spp. using partial rrs gene, 167 PCR products of expected size were obtained using primers specific to A. capra and 35 using those specific to A. platys. All these PCR products were sequenced. The 167 partial rrs gene sequences shared 99.3–99.7% nucleotide identities with those of A. capra, and the other 35 shared 99.1–99.6% nucleotide identities with those of A. platys and/or related strains complex. Co-infection with these two bacteria was found in 20 ticks. However, A. phagocytophilum and A. ovis were not detected in the analyzed ticks.
Phylogenetic relationship of the revealed A. platys-like bacteria with known strains
To better understand the phylogenetic relationships between the A. platys variants in this study and those described previously, the rrs gene sequences with the length of 1300 bp were amplified successfully from three representative positive samples. These three rrs gene sequences shared 100% nucleotide identities with each other, and 99.1–99.9% with known rrs gene sequences of specific A. platys, Candidatus A. camelii [34], and unclassified Anaplasma strains genetically related to A. platys (Additional file 2: Table S1). In the rrs tree, all these sequences (found in ticks, mosquitoes, dogs, goats, and cattle) had a close relationship with each other and formed a big cluster (named “specific A. platys and related strains”) separated from other species (Fig. 2a).
Phylogenetic position of revealed A. platys-like strains within Anaplasma genus. a Phylogenetic trees based on the rrs gene; (b) phylogenetic trees based on the gltA gene; (c) phylogenetic trees based on the groEL gene. Numbers at each node indicate bootstrap values. All three trees were rooted by mid-point methods. The scale bar represents the number of nucleotide substitutions per site. The taxa marked by circles depict the sequences obtained in this study
In addition to the rrs gene sequences, partial gltA gene sequences (about 600 bp) were recovered from all positive samples, and two nearly complete sequences (about 1150 bp) were recovered from the representative positive samples. For the partial gltA gene, it is notable that 34 strains (except AK-Rm383) shared 99.3–100% nucleotide identities and were distantly related to other known A. platys and related sequences, showing only 57.3–77.8% nucleotide identities (Additional file 2: Table S1). However, AK-Rm383 presented 96.3–97.9% nucleotide identities with variants identified in mosquitoes from Wuhan [35], and WHBMXZ-126 was also detected in R. microplus from Wuhan [36]. An uncultured Anaplasma sp. clone SY124 from Haemaphysalis longicornis tick in Shenyang [37] presented 52.6–60.3% nucleotide identities with other variants, including 34 strains recovered herein (Additional file 2: Table S1).
Consistent with the similarity analysis, all the specific A. platys and related strains were divided into three lineages in the phylogenetic tree of the gltA gene (Fig. 2b). All the specific A. platys strains formed a distinct lineage. The first lineage consisted of AK-Rm383, WHBMXZ-126, uncultured Anaplasma sp. clone SY124, and several sequences identified in mosquitoes from Wuhan. The second lineage included seven representatives from 34 strains obtained in the present study. In the gltA tree, the first distinct lineage was distant to Anaplasma sp. South Africa dog-1108 [38], and the uncultured Anaplasma sp. clone 1 and clone 2 [39]. The specific A. platys lineage was distant to the second one although they clustered together, sharing 74.0–77.2% identities. The position of Ca. A. camelii in the gltA-based tree was not determined due to the unavailability of its gltA gene.
In addition, partial groEL sequences (380 bp) were obtained from all positive samples, and five nearly complete sequences (about 1450 bp) were sequenced from selected positive samples. For the groEL gene, the partial sequences showed 99.7–100% nucleotide identities, and the nearly complete sequences presented 99.5–99.7% identities. In the groEL gene tree, all specific A. platys and related strains were divided into five lineages (Fig. 2c), which all clustered together. Furthermore, all eight representatives from 34 strains obtained in the present study (including AK-Rm383, WHBMXZ-126, Anaplasma sp. Mymensingh isolate BSH1 [40], several strains identified in mosquitoes from Wuhan, and two strains identified in dogs from the Philippines), clustered and formed the fourth lineage, whereas uncultured Anaplasma sp. clone SY124 clustered with HN238 and SHX21 rather than with the specific A. platys and related strains. All specific A. platys strains formed a distinct lineage, similar to that in the gltA tree. The first lineage composed by A. platys-like strains infecting Italian and Tunisian ruminants [41, 42] and Italian cats [43], as well as of A. platys-like strains detected in ticks from Italy [44] and Costa Rica [45]. The second lineage composed by uncultured Anaplasma sp. clones A1Bv10 and A1Bv6 detected in cattle from Tunisia, which were considered as A. platys-like strains by Ben Said et al. [41]. It is notable that Ca. A. camelii formed the third lineage and also fell into the diversity of the specific A. platys and related strains in the groEL tree, similar to that in the rrs tree. The nucleotide identities among specific A. platys and another four lineages of A. platys-like strains are shown in Table 3 and Table S1.
Phylogenetic relationship of A. capra in this study with known strains
Almost full-length rrs (about 1400 bp), and partial gltA (about 1100 bp) and groEL (about 1450 bp) DNA sequences were obtained from seven A. capra positive samples selected randomly. A comparison of the rrs gene sequences showed that all sequences had 99.6–100% nucleotide identity with each other and 99.0–100% nucleotide identities with known A. capra sequences (Additional file 2: Table S1). In the rrs tree, all revealed sequences clustered in the first lineage together with those of A. capra strains detected in goats, sheep, ticks, deer, and humans from China [3] and cattle (previously defined as A. centrale) from Japan (Fig. 3a). The second lineage included A. capra sequences detected in ticks, sheep, deer from China, and deer and serows from Japan.
Phylogenetic position of A. capra within Anaplasma genus. a Phylogenetic trees based on the rrs gene; (b) phylogenetic trees based on the gltA gene; (c) phylogenetic trees based on the groEL gene. Numbers at each node indicate bootstrap values. All three trees were rooted by mid-point methods. The scale bar represents the number of nucleotide substitutions per site. The taxa marked by circles depict the sequences obtained in this study
The topology of the partial groEL- and gltA-based phylogenetic trees resembled that of the rrs-based phylogenetic tree (Fig. 3b and c). In the trees based on the gltA and groEL genes, all revealed sequences clustered together in the first lineage with some sequences of A. capra strains detected in goats, sheep, ticks, giraffes (previously defined as A. phagoctophilum), humans from China, and cattle from Japan (previously defined as A. centrale). For both groEL and gltA genes, the nucleotide identities in the first lineage were 99.5–100% between the seven sequences in this study and 99.3–99.9% between the known sequences of A. capra (Additional file 2: Table S1).
Discussion
In the present study, we aimed to detect the Anaplasma species pathogenic to humans in ticks in Ankang city, Northwest China. Hard ticks, particularly Ixodes persulcatus and Haemaphysalis longicornis, are the main vectors that transmit Anaplasma species to humans and vertebrate animals by sucking blood [3, 9]. The primary vector for A. platys is considered to be R. sanguineus sensu lato [46,47,48]. The DNA of A. capra has been detected in R. microplus in Wuhan, China [36], whereas A. platys-like strains were also identified in R. microplus and R. turanicus [36, 49]. Here, R. microplus ticks were found infected with A. capra and A. platys-like strains by conventional PCR and sequencing. Co-infection by A. platys-like and A. capra was also observed. Hence, R. microplus may also be a vector for A. capra and A. platys-like strains, although further evidence is needed. Alternatively, these two pathogens may also be from animal hosts by sucking. Hence, their risk for humans is unknown and should be assessed in future studies.
A. capra was first detected in I. persulcatus in Heilongjiang Province, China, at 3% positive rate [3]. Subsequently, Sun et al. [9] identified it in H. longicornis in Shandong Province, China, at only 0.43% positive rate. Yang et al. [26] also found that A. capra-like strains circulated in H. qinghaiensis in Gansu Province, China, at 5.8% positive rate. Only 0.28% positive rate for A. capra was identified in R. microplus in Wuhan [36]. In our previous study, the positive rate was 40.4% for R. microplus and 22.6% for H. longicornis in Xi’an, China [29]. In this study, 36.6% (167/456) positive rate for A. capra was found in R. microplus ticks, similar to that for R. microplus and H. longicornis in Xi’an but higher than that for H. longicornis, H. qinghaiensis, I. persulcatus, and R. microplus in other previous studies [9, 26, 36]. A. platys-like strains were mainly detected in vertebrates rather than ticks. Only 1 of 354 R. microplus ticks in Wuhan was positive for A. platys-like strains [36], which is less than 7.68% found for R. microplus in this study.
Genetic and phylogenetic analysis based on the rrs, gltA, and groEL genes showed that specific A. platys strains formed a monophyletic group and shared more than 99.0% nucleotide identities regardless of whether they were collected from ticks, dogs, or other vertebrate animals, including humans. In recent years, several A. platys-like organisms were identified in cattle, sheep, goats, cats, and mosquitoes, sharing 86–93% similarity for the groEL gene with specific A. platys strains [35, 42, 43]. Furthermore, the gltA gene of A. platys-like strains from ticks and mosquitoes in China [35,36,37] formed a distinct lineage instead of clustering with specific A. platys strains (Fig. 2b). Another A. platys-like organism, detected in camels and named Ca. A. camelii [34, 50], clustered with specific A. platys strains in the rrs and groEL genes trees. In the groEL tree, A. platys-related strains detected in ticks from Ankang city had the closest relationship with strains recovered from mosquitoes [35], while only Ak-Rm383 collected in this study formed a distinct lineage in the gltA tree, clustering together with strains recovered from mosquitoes and ticks [35,36,37].
In this study, we used universal primers to obtain the groEL gene, and no sequencing results were ambiguous, which suggests that our tick samples were not co-infected with multiple strains of A. platys and related strains. Although we used primers specific to the gltA gene of the strains found in mosquitoes, we obtained only one sequence (data not shown), consistent with the results using the universal primers. Since the classification criteria for species of Anaplasma genus are not available, it is uncertain whether the lineages in the gltA and groEL phylogenetic trees, including Ca. A. camel, represent a distinct species or the same species.
In 2015, A. capra was considered as a new species and proved to be a human pathogen [3], although it has already been detected before in ticks and animals in China and Japan [37, 51,52,53]. After A. capra was considered to be pathogenic to humans, many molecular epidemiological investigations revealed its wide distribution in rural areas in China [9, 26, 35, 54]. The present study demonstrated the genetic diversity of A. capra. The first of two A. capra phylogenetic lineages was associated with the high positive rate in ticks. The strains described herein were most closely related to those detected in goats, sheep, various tick species, and humans based on the rrs, gltA, and groEL genes, revealing their genetic similarity. Hence, more attention should be paid to the infectivity of A. capra to humans.
The severity of the diseases caused by Anaplasma varies widely from mild (influenza-like illness) to severe (with lymphadenopathy and high hepatic aminotransferase concentrations). However, most cases caused by A. capra [3] and A. platys [5] mainly present as fever, headache, malaise, dizziness, and chills, and these nonspecific symptoms can easily be misdiagnosed as other infections [55]. A timely diagnosis of cases caused by A. capra is challenging for inexperienced clinicians, and laboratory identification is necessary under such circumstances. It is therefore urgent to develop rapid diagnostic procedures. Also, the pathogenicity of A. platys-like strains to humans should be determined in further studies.
Conclusions
A survey of Anaplasma species pathogenic to human was conducted in ticks collected from domestic animals in Ankang, China. Our results showed that A. capra and A. platys-like strains are present in R. microplus in the local area although their transmission to humans is uncertain. A. capra appears to be especially prevalent in ticks, and the strains identified in the present study are genetically similar to those previously detected in humans. Consequently, it should receive more attention from researchers and clinicians working at disease prevention and control. The pathogenicity of A. platys-like strains characterized in this study should also be assessed further.
Abbreviations
- HGA:
-
Human granulocytic anaplasmosis
- PCR:
-
polymerase chain reaction
References
Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, Ray SC, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51(Pt 6):2145–65.
Kocan KM, de la Fuente J, Cabezas-Cruz A. The genus Anaplasma: new challenges after reclassification. Rev Sci Tech. 2015;34(2):577–86.
Li H, Zheng YC, Ma L, Jia N, Jiang BG, Jiang RR, et al. Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect Dis. 2015;15(6):663–70.
Chen SM, Dumler JS, Bakken JS, Walker DH. Identification of a granulocy to tropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994;32(2):589–95.
Arraga-Alvarado CM, Qurollo BA, Parra OC, Berrueta MA, Hegarty BC, Breitschwerdt EB. Molecular evidence of Anaplasma platys infection in two women from Venezuela. Am J Trop Med Hyg. 2014;91(6):1161–5.
Chochlakis D, Ioannou I, Tselentis Y, Psaroulaki A. Human anaplasmosis and Anaplasma ovis variant. Emerg Infect Dis. 2010;16:1031–2.
Dugat T, Lagrée AC, Maillard R, Boulouis HJ, Haddad N. Opening the black box of Anaplasma phagocytophilum diversity: current situation and future perspectives. Front Cell Infect Microbiol. 2015;5:61.
Fang LQ, Liu K, Li XL, Liang S, Yang Y, Yao HW, et al. Emerging tick-borne infections in mainland China: an increasing public health threat. Lancet Infect Dis. 2015;15(12):1467–79.
Sun XF, Zhao L, Wen HL, Luo LM, Yu XJ. Anaplasma species in China. Lancet Infect Dis. 2015;15(11):1263–4.
Zhang L, Liu Y, Ni D, Li Q, Yu Y, Yu XJ, et al. Nosocomial transmission of human granulocytic anaplasmosis in China. JAMA. 2008;300(19):2263–70.
Wen B, Jian R, Zhang Y, Chen R. Simultaneous detection of Anaplasma marginale and a new Ehrlichia species closely related to Ehrlichia chaffeensis by sequence analyses of 16S ribosomal DNA in Boophilus microplus ticks from Tibet. J Clin Microbiol. 2002;40(9):3286–90.
Li Y, Chen Z, Liu Z, Liu J, Yang J, Li Q, et al. Molecular survey of Anaplasma and Ehrlichia of Red Deer and sika deer in Gansu, China in 2013. Transbound Emerg Dis. 2016;63(6):e228–36.
Yang J, Liu Z, Guan G, Liu Q, Li Y, Chen Z, et al. Prevalence of Anaplasma phagocytophilum in ruminants, rodents and ticks in Gansu, North-Western China. J Med Microbiol. 2013;62(Pt 2):254–8.
Yang J, Han R, Niu Q, Liu Z, Guan G, Liu G, et al. Occurrence of four Anaplasma species with veterinary and public health significance in sheep, northwestern China. Ticks Tick Borne Dis. 2018;9(1):82–5.
Kang YJ, Diao XN, Zhao GY, Chen MH, Xiong Y, Shi M, et al. Extensive diversity of Rickettsiales bacteria in two species of ticks from China and the evolution of the Rickettsiales. BMC Evol Biol. 2014;14:167.
Yang J, Li Y, Liu Z, Liu J, Niu Q, Ren Q, et al. Molecular detection and characterization of Anaplasma spp. in sheep and cattle from Xinjiang, Northwest China. Parasit Vectors. 2015;8:108.
Yu P, Liu Z, Niu Q, Yang J, Abdallah MO, Chen Z, et al. Molecular evidence of tick-borne pathogens in Hyalomma anatolicum ticks infesting cattle in Xinjiang Uygur autonomous region, Northwestern China. Exp Appl Acarol. 2017;73(2):269–81.
Yang Y, Cao J, Zhao H, Zhang J, Diwu J, Gao X. Investigation on species and nature geographic distribution of ticks in Shaanxi province. Chin J Hyg Insect & Equip Apr. 2008;14(2):97–9 (in chinese).
Chen Z, Li SS, Liu JZ. Illustrated keys to families and genera of the superfamily Ixodoidea under new taxonomic system. Chin J Parasitol Parasit Dis. 2011;29:302–4 (in Chinese).
Lu L, Wu HY. (2003). Classification and identification of important medical insects in China. Zhenzhou, Henan: Henan Science and Technology Press, 2003 (in Chinese).
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3(5):294–9.
Barlough JE, Madigan JE, DeRock E, Bigornia L. Nested polymerase chain reaction for detection of Ehrlichia equi genomic DNA in horses and ticks (Ixodes pacificus). Vet Parasitol. 1996;63:319–29.
Kawahara M, Rikihisa Y, Lin Q, Isogai E, Tahara K, Itagaki A, et al. Novel genetic variants of Anaplasma phagocytophilum, Anaplasma bovis, Anaplasma centrale, and a novel Ehrlichia sp. in wild deer and ticks on two major islands in Japan. Appl Environ Microbiol. 2006;72:1102–9.
Ochirkhuu N, Konnai S, Odbileg R, Murata S, Ohashi K. Molecular epidemiological survey and genetic characterization of Anaplasma species in Mongolian livestock. Vector Borne Zoonotic Dis. 2017;17:539–49.
Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703.
Yang J, Liu Z, Niu Q, Liu J, Han R, Liu G, et al. Molecular survey and characterization of a novel Anaplasma species closely related to Anaplasma capra in ticks, northwestern China. Parasit Vectors. 2016;9(1):603.
Martin AR, Brown GK, Dunstan RH, Roberts TK. Anaplasma platys: an improved PCR for its detection in dogs. Exp Parasitol. 2005;109:176–80.
Rar VA, Livanova NN, Panov VV, Doroschenko EK, Pukhovskaya NM, Vysochina NP, et al. Genetic diversity of Anaplasma and Ehrlichia in the Asian part of Russia. Ticks Tick Borne Dis. 2010;1(1):57–65.
Guo WP, Huang B, Zhao Q, Xu G, Liu B, Wang YH, et al. Human-pathogenic Anaplasma spp., and Rickettsia spp. in animals in Xi'an, China. PLoS Negl Trop Dis. 2018;12(11):e0006916.
Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–8.
Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–4.
Burland TG. DNASTAR’s Lasergene sequence analysis software. Methods Mol Biol. 2000;132:71–91.
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21.
Bastos AD, Mohammed OB, Bennett NC, Petevinos C, Alagaili AN. Molecular detection of novel Anaplasmataceae closely related to Anaplasma platys and Ehrlichia canis in the dromedary camel (Camelus dromedarius). Vet Microbiol. 2015;179(3–4):310–4.
Guo WP, Tian JH, Lin XD, Ni XB, Chen XP, Liao Y, et al. Extensive genetic diversity of Rickettsiales bacteria in multiple mosquito species. Sci Rep. 2016;6:38770.
Lu M, Tian JH, Yu B, Guo WP, Holmes EC, Zhang YZ. Extensive diversity of rickettsiales bacteria in ticks from Wuhan, China. Ticks Tick Borne Dis. 2017;8(4):574–80.
Dong X, Chen XP, Liu N, Dumler SJ, Zhang YZ. Co-circulation of multiple species of Rickettsiales bacteria in one single species of hard ticks in Shenyang, China. Ticks Tick Borne Dis. 2014;5(6):727–33.
Inokuma H, Oyamada M, Kelly PJ, Jacobson LA, Fournier PE, Itamoto K, et al. Molecular detection of a new Anaplasma species closely related to Anaplasma phagocytophilum in canine blood from South Africa. J Clin Microbiol. 2005;43(6):2934–7.
Ybañez AP, Matsumoto K, Kishimoto T, Inokuma H. Molecular analyses of a potentially novel Anaplasma species closely related to Anaplasma phagocytophilum detected in sika deer (Cervus nippon yesoensis) in Japan. Vet Microbiol. 2012;157(1–2):232–6.
Roy BC, Krücken J, Ahmed JS, Majumder S, Baumann MP, Clausen PH, et al. Molecular identification of tick-borne pathogens infecting cattle in Mymensingh district of Bangladesh reveals emerging species of Anaplasma and Babesia. Transbound Emerg Dis. 2018;65(2):e231–42.
Ben Said M, Belkahia H, El Mabrouk N, Saidani M, Alberti A, Zobba R, et al. Anaplasma platys-like strains in ruminants from Tunisia. Infect Genet Evol. 2017;49:226–33.
Zobba R, Anfossi AG, Pinna Parpaglia ML, Dore GM, Chessa B, Spezzigu A, et al. Molecular investigation and phylogeny of Anaplasma spp. in Mediterranean ruminants reveal the presence of neutrophil-tropic strains closely related to A. platys. Appl Environ Microbiol. 2014;80(1):271–80.
Zobba R, Anfossi AG, Visco S, Sotgiu F, Dedola C, Pinna Parpaglia ML, et al. Cell tropism and molecular epidemiology of Anaplasma platys-like strains in cats. Ticks Tick Borne Dis. 2015;6(3):272–80.
Chisu V, Zobba R, Lecis R, Sotgiu F, Masala G, Foxi C, et al. GroEL typing and phylogeny of Anaplasma species in ticks from domestic and wild vertebrates. Ticks Tick Borne Dis. 2018;9(1):31–6.
Campos-Calderón L, Ábrego-Sánchez L, Solórzano-Morales A, Alberti A, Tore G, Zobba R, et al. Molecular detection and identification of Rickettsiales pathogens in dog ticks from Costa Rica. Ticks Tick Borne Dis. 2016;7(6):1198–202.
Cicuttin GL, Tarragona EL, De Salvo MN, Mangold AJ, Nava S. Infection with Ehrlichia canis and Anaplasma platys (Rickettsiales: Anaplasmataceae) in two lineages of Rhipicephalus sanguineus sensu lato (Acari: Ixodidae) from Argentina. Ticks Tick Borne Dis. 2015;6(6):724–9.
Li Y, Yang J, Chen Z, Qin G, Li Y, Li Q, et al. Anaplasma infection of Bactrian camels (Camelus bactrianus) and ticks in Xinjiang, China. Parasit Vectors. 2015;8:313.
Simpson RM, Gaunt SD, Hair JA, Kocan KM, Henk WG, Casey HW. Evaluation of Rhipicephalus sanguineus as a potential biologic vector of Ehrlichia platys. Am J Vet Res. 1991;52(9):1537–41.
Harrus S, Perlman-Avrahami A, Mumcuoglu KY, Morick D, Eyal O, Baneth G. Molecular detection of Ehrlichia canis, Anaplasma bovis, Anaplasma platys, Candidatus Midichloria mitochondrii and Babesia canis vogeli in ticks from Israel. Clin Microbiol Infect. 2011;17(3):459–63.
Belkahia H, Ben Said M, Sayahi L, Alberti A, Messadi L. Detection of novel strains genetically related to Anaplasma platys in Tunisian one-humped camels (Camelus dromedarius). J Infect Dev Ctries. 2015;9(10):1117–25.
Liu Z, Ma M, Wang Z, Wang J, Peng Y, Li Y, et al. Molecular survey and genetic identification of Anaplasma species in goats from central and southern China. Appl Environ Microbiol. 2012;78(2):464–70.
Sato M, Nishizawa I, Fujihara M, Nishimura T, Matsubara K, Harasawa R. Phylogenetic analysis of the 16S rRNA gene of Anaplasma species detected from Japanese serows (Capricornis crispus). J Vet Med Sci. 2009;71(12):1677–9.
Zhou Z, Nie K, Tang C, Wang Z, Zhou R, Hu S, et al. Phylogenetic analysis of the genus Anaplasma in southwestern China based on 16S rRNA sequence. Res Vet Sci. 2010;89(2):262–5.
Yang J, Liu Z, Niu Q, Liu J, Han R, Guan G, et al. A novel zoonotic Anaplasma species is prevalent in small ruminants: potential public health implications. Parasit Vectors. 2017;10(1):264.
Dahlgren FS, Heitman KN, Behravesh CB. Undetermined human Ehrlichiosis and Anaplasmosis in the United States, 2008-2012: a catch-all for passive surveillance. Am J Trop Med Hyg. 2016;94(2):299–301.
Acknowledgements
Not applicable.
Funding
This study was funded by the National Natural Science Foundation of China (No. 31700159). The funders had no role in study design, data collection and analysis, or preparation of the manuscript.
Availability of data and materials
The rrs, gltA and groEL genes sequences generated in this study were submitted to the GenBank database under the accession numbers MH716407-MH716436 and MH762071- MH762085.
Author information
Authors and Affiliations
Contributions
WPG and EMZ conceived and designed the study and critically revised the manuscript. WPG, BZ and XW performed tick collection. WPG, BZ, YHW, GX and XN conducted the laboratory experiments. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
An ethical approval was obtained from the Institutional Animal Care and Use Ethics Committee of Northwest A&F University, Northwest A&F University. Verbal consent was obtained from the owners of animals for tick samples to be taken.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Figure S1. The partial gltA and groEL genes of A. platys-like strains and different fragments of partial and nearly complete rrs, gltA, and groEL genes of A. platys-like strains and A. capra amplified with different pairs of primers from the tick DNA specimens. (TIF 360 kb)
Additional file 2:
Table S1. Information for the sequences submitted to GenBank database used for phylogenetic analysis. (DOCX 18 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Guo, WP., Zhang, B., Wang, YH. et al. Molecular identification and characterization of Anaplasma capra and Anaplasma platys-like in Rhipicephalus microplus in Ankang, Northwest China. BMC Infect Dis 19, 434 (2019). https://doi.org/10.1186/s12879-019-4075-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12879-019-4075-3