Abstract
Background
β-Glucosidases are components of the cellulase system, a family of enzymes that hydrolyze the β-1,4 linkages of cellulose. These proteins have been extensively studied due to the possibility of their use in various biotechnological processes. They have different affinities for substrates (depending on their source) and their activities can be used for saccharification of different types of biomass. In this context, the properties and the synergistic capacity of β-glucosidases from different organisms, to supplement the available commercial cellulase cocktails, need a comprehensive evaluation.
Results
Two β-glucosidases belonging to GH3 family were secreted by Penicillium citrinum UFV. PcβGlu1 (241 kDa) and PcβGlu2 (95 kDa) presented acidic and thermo-tolerant characteristics. PcβGlu1 showed Michaelis–Menten kinetics for all substrates tested with Km values ranging from 0.09 ± 0.01 (laminarin) to 1.7 ± 0.1 mM (cellobiose, C2) and kcat values ranging from 0.143 ± 0.005 (laminarin) to 8.0 ± 0.2 s−1 (laminaribiose, Lb). PcβGlu2 showed substrate inhibition for 4-methylumbelliferyl-β-d-glucopyranoside (MUβGlu), p-nitrophenyl-β-d-glucopyranoside (pNPβGlu), cellodextrins (C3, C4, and C5), N-octil-β-d-glucopyranoside, and laminaribiose, with Km values ranging from 0.014 ± 0.001 (MUβGlu) to 0.64 ± 0.06 mM (C2) and kcat values ranging from 0.49 ± 0.01 (gentiobiose) to 1.5 ± 0.2 s−1 (C4). Inhibition constants (Ki) for PcβGlu2 substrate inhibition ranged from 0.69 ± 0.07 (MUβGlu) to 10 ± 1 mM (Lb). Glucose and cellobiose are competitive inhibitors of PcβGlu1 and PcβGlu2 when pNPβGlu is used as a substrate. For PcβGlu1 inhibition, Ki = 1.89 ± 0.08 mM (glucose) and Ki = 3.8 ± 0.1 mM (cellobiose); for PcβGlu2, Ki = 0.83 ± 0.05 mM (glucose) and Ki = 0.95 ± 0.07 mM (cellobiose). The enzymes were tested for saccharification of different biomasses, individually or supplementing a Trichoderma reesei commercial cellulose preparation. PcβGlu2 was able to hydrolyze banana pseudostem and coconut fiber with the same efficiency as the T. reesei cocktail, showing significant synergistic properties with T. reesei enzymes in the hydrolysis of these alternative biomasses.
Conclusions
The β-glucosidases from P. citrinum UFV1 present different enzymatic properties from each other and might have potential application in several biotechnological processes, such as hydrolysis of different types of biomass.
Similar content being viewed by others
Background
β-Glucosidases (EC 3.2.1.21) constitute a group of well-characterized enzymes, displaying several functions, of biological and industrial relevance. These enzymes catalyze the hydrolysis from the nonreducing termini of β-glycosidic bonds present in short-chain oligosaccharides (containing 2–6 monosaccharides), alkyl- and aryl β-d-glucosides [1, 2]. They are also involved in transglycosylation reactions of β-glucosidic linkages of glucose conjugates. β-Glucosidases are widely distributed in living organisms, and the affinity of these enzymes for a particular substrate is dependent on their physiological function, location, and the nature of the enzyme source [3].
β-Glucosidases are also a component of the cellulase system, a family of enzymes that hydrolyze the β-1,4 linkages of cellulose, which presents three categories of enzymatic components: (1) endo-glucanases (EC 3.2.1.4), which act on the cellulose chain catalyzing the random cleavage of internal bonds to yield glucose and cello-oligosaccharides; (2) cellobiohydrolases (EC 3.2.1.91), which release cellobiose from the reducing and nonreducing ends of the polysaccharide, and (3) β-glucosidases (cellobiase, EC 3.2.1.21), which release glucose from cello-oligosaccharides [4, 5]. Because both endoglucanase and exoglucanase activities are usually inhibited by cellobiose and short cello-oligosaccharides, β-glucosidases are responsible for the rate-limiting step of the whole cellulolytic process in vitro [6, 7].
These enzymes constitute a significant group among glycoside hydrolases, and the possibility of their use in various biotechnological processes has been intensely explored. Considering industrial applications, β-glucosidases are currently used in: production of biodegradable nonionic surfactants and other compounds [8]; synthesis of diverse oligosaccharides, glycoconjugates, alkyl- and amino-glycosides [5]; detoxification of cassava [9]; removal of cyanogenic glucosides from sorghum malt used in the production of African beer [10]; enzymatic release of aroma compounds from glucosidic precursors present in fruit juices during winemaking [11]; enhancement of tea extracts aroma [12], among others. These enzymes can play a critical role in the generation of potentially sustainable energy sources (e.g., glucose, ethanol, hydrogen, and methane) from biomass conversion [13].
The production of ethanol from renewable sources has gained attention in the last decade due to the concern over depleting fossil fuel and the impact on the environment. Different bio-renewable materials have arisen as potential sources for alternative fuel production [14], such as sugars, starch, and lignocellulosic material. The lignocellulosic biomass is abundantly available and is a ubiquitous source of energy; most importantly, it does not compete with food production and animal feed. Cellulosic sources as corn stover, sugarcane bagasse, rice, and wheat straws are the most promising sources to be uses as substrates for bioethanol production [15,16,17,18]. Moreover, this other lignocellulosic materials can be obtained from industrial wastes, wood, and agricultural residues.
Commercial cellulolytic preparations for biomass hydrolysis, such as Celluclast ® (Novo Nordisk), Acellerase® (Sigma-Aldrich), and the newly developed Cellic® CTec2 and Cellic® CTec3 (Novozymes), incorporate fungal glycosyl hydrolases in their composition, especially from Trichoderma reesei. One of the most studied fungi for cellulose hydrolysis is Trichoderma reesei, as it secretes a cellulase mixture composed mainly of exoglucanases and endo-glucanases (> 92%), albeit just a few β-glucosidases [19]. Hence, an efficient biomass conversion of commercial preparations using T. reesei is dependent on the supplementation with exogenous β-glucosidase. High amounts of cellobiose are produced by exo- and endo-glucanases, but β-glucosidases undertake the rate-limiting step by hydrolyzing cellobiose to glucose [20]. In short, these are crucial enzymes for the process of biomass bioconversion, and finding a β-glucosidase capable of efficiently supplementing these cocktails remains a major bottleneck, since cellobiose is a potent inhibitor of cellulase activities.
A few fungal strains are known to be efficient producers of β-glucosidases. Among them, some filamentous thermophilic fungi are good sources of β-glucosidases with high thermal stability, a most desirable property for industrial purposes [21]. Several works have demonstrated the production of β-glucosidases from different fungal species, including Chrysoporthe cubensis [22], Talaromyces leycettanus [20], Acremonium thermophilum (AtBG3), and Thermoascus aurantiacus (TaBG3) [23], aiming supplementation to enhance the saccharification efficiency of cellulosic materials.
In this study, we isolated a cellulolytic fungus, Penicillium citrinum UFV1, from sugarcane bagasse. Production of an endoglucanase, xylanase, and β-galactanase [24,25,26] from P. citrinum has been reported and β-glucosidase activity from Penicillium citrinum YS40-5 was already described [4]. However, the detailed kinetic properties and the possible application of these enzymes for the saccharification process of natural biomass were not yet explored.
Exploiting the fact that these enzymes have different affinities for substrates, depending on enzyme source, the present study describes (a) the production, purification, and biochemical characterization of two β-glucosidases from P. citrinum UFV1; (b) these enzymes’ application on the saccharification process of sugar cane bagasse and two other alternative biomasses, coconut fiber and colloidal banana pseudostem; and (c) their synergistic effect to the Trichoderma reesei cellulase system (the most common commercial source of cellulases).
Results
Enzyme production
First, we investigated the potential of P. citrinum UFV1 to produce β-glucosidases growing on low-cost biomass. The fungus was grown for 10 days under submerged culture using wheat bran as a carbon source and β-glucosidase activity in the soluble fraction was monitored (Fig. 1). β-Glucosidase activity increased until 8–9 days of fermentation when maximal specific activities were observed (4.3 ± 0.3 - 4.4 ± 0.1 U mg−1). Other glycoside hydrolases were produced in these conditions, especially xylanase and cellulase (Additional file 1: Table S1). A fermentation lasting 8 days was established as the standard protocol for enzyme production, and subsequent purification of P. citrinum UFV1 β-glucosidases.
Purification and sequence identification by mass spectrometry
Penicillium citrinum UFV1 β-glucosidases were successfully purified in few steps (Table 1). The supernatant culture was subjected to 50% ammonium sulfate precipitation and the remaining supernatant, after centrifugation, was applied to a hydrophobic interaction chromatography. The activity was eluted in two peaks after chromatography, PcβGlu1 and PcβGlu2 (Fig. 2a). PcβGlu1 was more active on pNPβGlu than on MUβGlu, while PcβGlu2 was more active on MUβGlu (Fig. 2a). After this step, PcβGlu1 was obtained as a homogeneous protein (see below). For further purification of PcβGlu2 and molecular mass measurements, PcβGlu1 and PcβGlu2 were applied onto a gel filtration Superdex S200 column (AKTA Purifier); each active enzyme displayed its own retention time (Fig. 2b, c). PcβGlu1 was purified 3.9-fold with a yield of about 9.9%, while PcβGlu2 was purified 3.4-fold with a yield of 3.2% (Table 1).
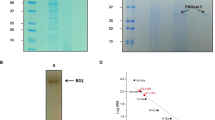
Chromatographic profile obtained after β-glucosidase purification. a Pellet from ammonium sulfate precipitation was resuspended and applied onto a phenyl sepharose column (AKTA PRIME, GE Healthcare Biosciences). Purified PcβGlu1—fractions 12–16. PcβGlu2—fractions 25–30. Activity was determined using pNPβGlu and MUβGlu substrates. b PcβGlu1 fractions from phenyl sepharose were pooled and applied onto a Superdex 200 column for molecular mass determination. c PcβGlu2 fractions from phenyl sepharose were pooled and applied onto a Superdex 200 column fractions for further purification and molecular mass determination. Purified PcβGlu2—fractions 29–32. Activity was measured with MUβGlu substrate
Purified PcβGlu1 and PcβGlu2 were revealed as unique bands when submitted either to PAGE (detection by in-gel assays with MUβGlu) or SDS-PAGE (protein staining) indicating the homogeneity of the preparations (Fig. 3, lanes 2–5). Besides, PcβGlu1 and PcβGlu2 activity bands corresponded to the two regions of activity observed in the soluble fraction of P. citrinum UFV1 (Fig. 3, lanes 6–9).
SDS-PAGE and in-gel assays after PAGE of PcβGlu1 and PcβGlu2. Lanes 1–5: SDS-PAGE (silver staining); lanes 6–9: in-gel assays using MUβGlu as a substrate. 1 Molecular mass markers. 2 and 6 P. citrinum crude extract. 3 and 7 PcβGlu1 after phenyl sepharose (see Fig. 2a). 4 and 8 PcβGlu2 after phenyl sepharose (see Fig. 2a). 5 and 9 PcβGlu2 after chromatography in Superdex 200 column (see Fig. 2c)
The molecular mass of PcβGlu1 was estimated as 134 kDa by SDS-PAGE (Fig. 3, lanes 3) and 258 kDa by gel filtration chromatography (Fig. 2b). Hence, it was assumed that native PcβGlu1 is a homodimeric protein. Analysis of PcβGlu2 using the same techniques and conditions revealed a molecular mass of 93 kDa (SDS-PAGE) and 89 kDa (gel filtration chromatography), suggesting that the native form of this protein is monomeric.
Purified PcβGlu2 was very unstable after removal of the ammonium sulfate from the chromatographic buffer (data not shown). The addition of substrates, sugars, detergents, and proteins was tested for stabilization at − 20 and 4 °C (Additional file 2: Table S2). The best stabilizer was bovine albumin serum (BSA) at 1 mg mL−1, which maintained PcβGlu2 activity for at least 1 week at 4 °C.
Purified PcβGlu1 was characterized by shotgun mass spectrometry coupled to data analysis with the PEAKS Studio software (http://www.bioinfor.com/peaks-studio/) against a comprehensive Uniprot “Penicillium” database (338,849 entries). Identified peptides matched several protein groups, with a major contribution of sequences from the glycoside hydrolase family 3 (GH3) (Additional file 3: Table S3). It is important to notice that none of the other hits obtained corresponded to protein families with β-glucosidase activity (E.C. 3.2.1.21), with some of these sequences matching pseudogenes or protein entries with no associated catalytic activity information. Unfortunately, analysis of purified Pcβglu2 did not yield significant results, mostly due to sample contamination with keratin (data not shown). Hence, we present here the data that we had previously obtained using the heterogeneous fraction obtained following hydrophobic interaction chromatography (Fig. 2a), which was enriched for this enzyme. As expected, the results showed a high diversity of protein contaminants, but we observed a sizable contribution of protein sequence hits belonging to the GH3 family (Additional file 4: Table S4). Despite the identification of an array of glycoside hydrolases from different families, all the protein sequences identified in this sample that do not belong to GH3 lack both beta-glucosidase activity and the catalytic active site residues (as the proton donor and nucleophile) that are typical for this enzyme group (Additional file 3: Table S3, Additional file 4: Table S4). In this respect, mass spectrometry data indicate that both PcβGlu1 and PcβGlu2 might belong to the GH3 group of enzymes, with a more confident identification for the first enzyme.
Effect of pH, temperature, and metal ions on PcβGlu1 and PcβGlu2
Effect of pH on the β-glucosidases activities was determined with MUβGlu as a substrate at 40 °C and different pH (3–10). For pH stability evaluation, the enzymes were incubated at 40 °C for 4 h at various pH before residual activity determination. Both enzymes were stable for 4 h at 40 °C at pH range 5–8 (Fig. 4). Purified PcβGlu1 and PcβGlu2 showed maximal activity at pH 5 (Fig. 4a) and 5–6 (Fig. 4b), respectively. PcβGlu1 was more susceptible to pH change than PcβGlu2.
Effect of pH on PcβGlu1 and PcβGlu2 activities. Enzymes were incubated in different buffers from pH 3 to 10 at 40 °C. For stability tests, the enzymes were pre-incubated for 4 h and residual activity was measured at the optimum pH. Activities were measured with MUβGlu as a substrate. a PcβGlu1. b PcβGlu2
Thermal stability of β-glucosidases was determined by incubating the enzymes for 4 h at 30–80 °C before residual activity determination. PcβGlu1 was stable in the presence of ammonium sulfate during 4 h at 50, 60, and 70 °C; at 80 °C, the enzyme was inactivated after 15 min (Fig. 5a). In the absence of ammonium sulfate, PcβGlu1 showed a half-life of 51 ± 2 min and kd of 2.3 × 10−4 ± 1 × 10−5 s−1 at 50 °C.
PcβGlu2 was stable in the presence of BSA (1 mg mL−1) for 4 h at 30 and 40 °C (Fig. 5b). For 50 °C and 60 °C, the calculated half-life is demonstrated in Table 2. At 70 °C, the enzyme was completely inactivated after 4 min (data not show). In the absence of BSA, PcβGlu2 showed a half-life of 36 ± 6 min and kd of 3.4 × 10−4 ± 5 × 10−5 s−1 at 30 °C. Values of kd and half-life for PcβGlu1 and PcβGlu2 are demonstrated in Table 2; it is important to stress that kd values are inversely proportional to the protein’s thermostability, that is, the higher the kd, the lower will be the protein stability. Hence, the data confirm (Table 2) that PcβGlu1 was more thermostable than PcβGlu2.
As shown in Fig. 5, the increase in temperature resulted in a progressive inactivation of both enzymes, PcβGlu1 being more thermo-tolerant than PcβGlu2. The thermostability of PcβGlu1 is increased by the presence of ammonium sulfate. At low concentrations, this salt can stabilize proteins by non-specific electrostatic interactions which depend on the ionic strength of the medium [27]. PcβGlu1 and PcβGlu2 activity loss at high temperatures followed first-order kinetics, which indicated an irreversible inactivation by a monomolecular process [28].
Effects of various ions on β-glucosidases activity were investigated by pre-incubating the enzymes for 30 min at 40 °C in the presence of these ions before residual activity determination. Purified PcβGlu1 and PcβGlu2 activities were highly affected in the presence of 100 mM Fe3+, Ba2+, and Pb2+ (Table 3). PcβGlu2 was also affected by Cu2+ and Hg+, whereas PcβGlu1 showed higher residual activities of 29.6 ± 0.4 and 56 ± 3% after incubation with Cu2+ and Hg+, respectively. No significant inhibition occurred in the presence of Ca2+, Zn2+, Co2+, and Mn2+ for PcβGlu1. Nevertheless, PcβGlu2 activity was affected especially by Zn2+ incubation. Small activation of PcβGlu1 and inhibition of PcβGlu2 was observed in the presence of Mg2+.
Kinetic parameters and subsite analysis
Kinetic parameters of purified β-glucosidases were determined using series of concentrations of different substrates. PcβGlu1 and PcβGlu2 showed broad substrate specificity, hydrolyzing pNPβGlu, MUβGlu, OctylβGlu, cellobiose, cellotriose, cellotetraose, cellopentaose, gentiobiose, laminaribiose, and laminarin (Tables 4, 5). In contrast, both enzymes showed no hydrolytic activity against pNPβGal, MUβMan, Avicel®, or CMC (data not shown). The configuration of the anomeric carbon of released glucose molecules by PcβGlu1 and PcβGlu2 is β (data not show), as determined by the glucose oxidase method (see “Methods”). This result suggested that the reaction catalyzed by these enzymes occured by the classical mechanism of retention of configuration of the anomeric carbon.
Kinetic parameters for the hydrolysis of various substrates by purified PcβGlu1 and PcβGlu2 are presented in Tables 4 and 5, respectively.
The best substrates for PcβGlu1 were pNPβGlu and MUβGlu, with higher catalytic efficiencies values. PcβGlu2 hydrolyzed MUβGlu more efficiently than PcβGlu1, which preferred pNPβGlu as a substrate. Differences in catalytic efficiencies of PcβGlu1 against these substrates were due mainly to variations in the kcat value, whereas for Pcβglu2, these differences resulted from different binding affinities of the enzyme to the substrates (e.g., lower Km for MUβGlu).
For cellodextrins, the highest catalytic efficiency was identified for cellopentaose hydrolysis by PcβGlu1 and PcβGlu2. PcβGlu1 hydrolyzes laminaribiose, gentiobiose, and octilβGlu with similar catalytic efficiencies, with a lower efficiency on laminarin (5.9 ± 0.9 s−1 mM−1). PcβGlu2 showed a high catalytic efficiency against laminarin (44.3 ± 1.8 s−1 mM−1), mainly due to a high binding affinity (lower Km) to this substrate. PcβGlu2 showed very low catalytic efficiencies for laminaribiose, octylβGlu, and gentiobiose.
PcβGlu1 showed classical Michaelis–Menten kinetics for all substrates. However, PcβGlu2 showed classical Michaelis–Menten kinetics when acting on gentiobiose, cellobiose, and laminarin hydrolysis, and showed enzyme inhibition by high concentrations of the substrates pNPβGlu, MUβGlu, laminaribiose, cellotriose, cellotetraose, cellopentaose, and octylβGlu (Fig. 6a–c). It is interesting to note the results obtained for cellodextrins, where cellotetraose showed the lowest Ki value (1.1 ± 0.3 mM), inhibiting PcβGlu2 at a lower concentration than cellotriose and cellopentaose (Table 5).
Effect of substrate concentration on PcβGlu2 velocity. Theoretical curves (lines) were determined using kinetic parameters from Table 5. a 4-Methylumbelliferyl β-d-glucopyranoside and ρ-nitrophenyl-glucopyranoside. b cellotriose, cellotetraose, and cellopentaose. c laminaribiose and octyl-β-glucopyranoside
Glucose competitively inhibited the hydrolysis of pNPβGlu by PcβGlu1 and PcβGlu2, with Ki values of 1.89 ± 0.08 and 0.88 ± 0.08 mM, respectively (Fig. 7a, b). Cellobiose inhibited the hydrolysis of pNPβGlu by PcβGlu1 and PcβGlu2 in a mixed-type noncompetitive mode, with KIE = 10.7 ± 0.1 mM and KIES = 18.7 mM for PcβGlu1 (Fig. 7c) and KIE = 3.74 ± 0.07 mM and KIES = 6.2 mM for PcβGlu2 (Fig. 7d). These values reflect a higher affinity binding to the inhibitor for the free enzyme (KIE) than for the enzyme–substrate complex (KIES).
Graphic analysis of the inhibition of P. citrinum β-glucosidases. Lineweaver–Burk plot of initial velocity versus various fixed substrate concentrations showing inhibitory effects of glucose on pNPβGlu hydrolysis by a PcβGlu1 and b PcβGlu2. Inset: slope plot from the same data. Lineweaver–Burk plot of initial velocity versus various fixed substrate concentrations showing inhibitory effects of cellobiose on pNPβGlu hydrolysis by c PcβGlu1 and d PcβGlu2. Insets: slope and intercept plots from the same data
There were at least five subsites for binding glucosyl residues in the active sites of PcβGlu1 and PcβGlu2, numbered − 1, + 1, + 2, + 3, and + 4 (Fig. 8). The substrate is hydrolyzed between subsites − 1 and + 1. The subsite + 1 and subsite + 2 had higher affinities for glucosyl residues than the other subsites in both enzymes (Fig. 8), but the profile observed demonstrated differences in the catalytic site of these enzymes. The intrinsic catalytic constant was determined to be 49.2 s−1 for PcβGlu1 and 9.4 s−1 for PcβGlu2. The maximum deviation within 14% between experimental and theoretical rate parameters confirmed the validity of the equations and the calculated subsite affinities (Additional file 5: Table S5).
Biomass saccharification
We supplemented the cellulases from T. reesei with purified PcβGlu1 and PcβGlu2 from P. citrinum UFV1 for hydrolysis of colloidal preparations of sugarcane bagasse, banana leaf, and coconut fiber with minimal pre-treatment [30]. The enzymes of P. citrinum UFV1 were able to hydrolyze coconut fiber with the same capacity as T. reesei cellulases (Fig. 9c, f), and the quantity of reducing sugars released from this source of biomass after 24 h of saccharification was increased when T. reesei cocktail was supplemented with PcβGlu2 (Table 6).
Time course of glucose production in experiments of enzymatic saccharification of colloidal biomasses (2.5% (w/v)) by T. reesei cellulases and PcβGlu1 or PcβGlu2. a, d Hydrolysis of sugar cane. b, e Hydrolysis of banana pseudostem. c, f Hydrolysis of coconut fiber. a–c Supplementation of T. reesei cellulases with PcβGlu1, and d–f show supplementation with PcβGlu2. Ten CBU/g of biomass were used for PcβGlu1 and PcβGlu2, while for T. reesei cellulases 7 CBU/g were used
However, while PcβGlu2 was able to hydrolyze sugarcane bagasse (2.5% maximum hydrolysis) and banana pseudostem (10.5% maximum hydrolysis) (Fig. 9d, e), PcβGlu1 had no activity on these substrates (Fig. 9a, b). When using banana pseudostem as a substrate, Pcβglu2 had the same efficiency as T. reesei enzymes and demonstrated the ability to supplement the cellulases of T. reesei in banana pseudostem hydrolysis. The maximum percentage of hydrolysis of this substrate, after 24 h of saccharification, increased from 10.6 to 18.0% with the addition of PcβGlu2, and the amount of reducing sugars generated increased from 11.9 to 20.0 mM (Fig. 9e and Table 6).
It is interesting to note that the T. reesei cocktail displayed different reducing sugar production capacities depending on the type of biomasses. For instance, this enzyme sources had better activity on sugarcane bagasse than on coconut fiber.
The results demonstrated that PcβGlu1 and PcβGlu2 have different abilities to hydrolyze colloidal substrates (Table 6) and that PcβGlu2 is the most promising for supplementation of T. reesei cellulases aiming the hydrolysis of biomass.
Discussion
Filamentous fungi are known as good producers of β-glucosidases and a large number of β-glucosidases from family GH1 and GH3 have been purified and characterized from these organisms [31]. The ability of fungi to grow on low-cost substrates makes them potential sources of enzymes for industrial applications. The production of cellulases and hemicellulases by filamentous fungi is strongly affected by culture conditions. The carbon source plays a central role in enzyme production, and cellulolytic enzymes are better induced by carbohydrates or their derivatives [32]. In this study, two β-glucosidases with different properties (PcβGlu1 and Pcβglu2) were produced and purified from Penicillium citrinum UFV1 cultivated in submerged culture, using wheat bran as carbon source.
The results for β-glucosidase production by P. citrinum UFV1 cultivation in submerged fermentation (SMF), containing wheat bran as a carbon source, have the same order of magnitude than those obtained by well-known cellulolytic species, such as Aspergillus spp., Penicillium spp., and Trichoderma spp., which are employed for the industrial production of cellulases [33,34,35]. Several investigations with different cellulolytic microorganisms have emphasized the advantages of wheat bran as a substrate to obtain high cellulase productions [33, 36]. Besides, wheat bran is a byproduct of industries and a carbon source broadly available.
The production of β-glucosidases isoforms by filamentous fungi has been previously reported [5, 37, 38] and the multiplicity of β-glucosidases can be attributed to the presence of multiple genes or due to differential post-transcriptional modifications [39]. As we demonstrated in this work, two different β-glucosidases were identified for Penicillium citrinum in the conditions studied. The purified PcβGlu1 and PcβGlu2 had different molecular masses, being characterized as homodimeric and monomeric proteins, respectively. Following peptide identification by mass spectrometry, in conjunction with available literature data and purified PcβGlu1/PcβGlu2 enzymatic characterization (this work), both enzymes were tentatively classified as belonging to the GH3 family. Dimeric β-glucosidases were described for Paecilomyces thermophile [21] and Humicola insolens [37], showing native molecular masses of about 200 kDa. Values ranging from 40 to 250 kDa were reported for β-glucosidases from different fungal sources, with wide variation concerning their molecular mass and the homogeneity of subunits, mainly due to post-translational modifications, since some β-glucosidases are highly glycosylated [3].
The structure of GH3 members includes an N-terminal (α/β)8 barrel followed by a C-terminal (α/β)6 sandwich domain. In general, the catalytic nucleophile is an aspartate found in the conserved motif VMSDW located in the TIM barrel domain and a glutamate acts as a proton donor in the catalytic site [40]. The active site is situated in the pocket at the interface between the two domains [1, 3, 41]. Additional domains can be found in some GH3 structures like the C-terminal fibronectin type III domain in Bgl3B from T. neapolitana and also in KmBglI from K. marxianus. The latter also has a PA14 domain inserted into the (α/β)6 sandwich domain that influences carbohydrate-binding [42, 43].
The classification of PcβGlu1 and PcβGlu2 in the GH3 family is consistent with the results showing that a retention mechanism exists and that Cu2+ and Zn2+ were inhibitory to the enzymatic activity of these proteins, which further supports the notion that basic (Arg, Lys, and His) and acidic (Asp and Glu) amino acids can be present in the catalytic domains [44]. The fact that these enzymes are differently affected by these metal ions demonstrates differences in their catalytic domains.
The determinations of enzymatic characteristics showed that PcβGlu1 and PcβGlu2 are acidic β-glucosidases with stability in a pH range from 5 to 8 and that PcβGlu1 is more thermostable than PcβGlu2. PcβGlu1 showed an estimated half-life of 150 h at 70 °C and PcβGlu2 of 13 h at 50 °C. The thermostability of these enzymes is an interesting characteristic for application in various biotechnological processes. In general, β-glucosidases produced by different fungi species have an acidic characteristic with highest activities between pH 4 and 6 [34, 46, 47]; the thermal stability of these enzymes is comparable to other thermo-tolerant β-glucosidases reported [2, 4, 20, 47]. However, it is important to highlight that our results suggest that PcβGlu1 and Pc βGlu2 require specific conditions for stabilization, such moderate ionic strength (obtained with ammonium sulfate in our experiments) or moderate protein concentrations (mimicked with BSA in our tests). Both restraints should probably not be impeditive their application in industry, as these conditions are normally obtained in general biomass degradation reactions.
Based on substrate specificity, β-glucosidases have been classified as (i) aryl β-glucosidases, which act on aryl-glucosides, (ii) true cellobiases, which hydrolyze cellobiose to release glucose, and (iii) broad substrate specificity β-glucosidases, which act on a wide spectrum of substrates [3]. PcβGlu1 and PcβGlu2 from P. citrinum UFV1 showed significant activity for both aryl-glucosides and cello-oligosaccharides, indicating that they belong to the group of broad substrate specificity β-glucosidases. Moreover, PcβGlu2 hydrolyzed laminarin with high efficiency, indicating that this β-glucosidase can display an additional exo-β-1,3-glucanase activity. Both glucosidases hydrolyzed β-1,3, β-1,4 and β-1,6 linked disaccharides. Broad specificity and exo-β-glucanase activity were already described for other GH3 glucosidases [45, 48, 49].
Kinetic parameters for various β-glucosidases have been determined using different substrates in the presence and the absence of glucose and Ki and Km values have a lot of variation [23, 50]. PcβGlu1 and PcβGlu2 Km values decreased with the increasing length of the substrates cellobiose, cellotriose, cellotetraose, and cellopentaose, suggesting that these enzymes have at least five subsites which bind glucosyl residues. An extended binding site also suggests a role for these enzymes in the degradation of oligosaccharides beyond the hydrolysis of cellobiose and other disaccharides, reinforcing their broad specificity. As the intrinsic catalytic constant calculated for PcβGlu1 (49.2 s−1) and PcβGlu2 (9.43 s−1) are higher than the catalytic constants determined for cellodextrins (Table 6), non-productive complexes are expected to be formed [29].
For Pcβglu2, some substrates did not follow classical Michaelis–Menten kinetics, because enzyme was inhibited by high concentrations of substrate (Fig. 6). This fact can be explained by transglycosylation reactions or by the binding of a second molecule of the substrate in the active site (substrate inhibition). Different works have found that for GH3 β-glucosidases with a retaining mechanism, the reduced hydrolytic activity relies at least in part on the occurrence of transglycosylation [38, 45, 51, 52]. Coherently, PcβGlu1 and PcβGlu2 are retaining enzymes, and β-glucosidases that hydrolyze substrates by double displacement mechanisms are classified in families GH1, GH3, GH5, GH16, and GH30 [53]; this kind of mechanism also permitting enzymes to transglycosylate.
Glucose and cellobiose inhibited PcβGlu1 and PcβGlu2 in competitive and mixed noncompetitive modes of inhibition, respectively. These types of inhibition suggest a second binding site for cellobiose in the active site of these enzymes near the active site, as the Ki values determined for cellobiose are higher than its Km values. This second binding site could correspond to peripheral subsites in the active site, but more evidence is necessary to corroborate this hypothesis.
The β-glucosidases studied in this work showed different specificities and characteristics and they are potential sources to be used in different biotechnological applications. The rate and cost limiting steps in the production of biofuels from biomass in the conversion of polysaccharides to glucose are well known [20]. Consequently, β-glucosidases acting synergically with other enzymes could improve the results of biomass transformation making the process viable. We supplemented the cellulases from T. reesei with purified PcβGlu1 and PcβGlu2 from P. citrinum UFV1 for hydrolysis of colloidal preparations of sugarcane bagasse, banana leaf, and coconut fiber with minimal pre-treatment [30]. The supplementation assessed the synergistic effect of P. citrinum UFV1 PcβGlu2 on T. reesei cellulases activities.
Abundantly available lignocellulosic crop residues as sugar cane bagasse, coconut fiber or banana pseudostem can be used in biorefineries for ethanol production. Some studies have shown that different biomasses have different chemical compositions [54, 55]. Hence, cellulolytic enzymes secreted by different organisms, and induced in liquid or solid medium, can act differently on these natural substrates. Specificities of the enzymes will determine the type of material they can act upon efficiently.
The colloidal biomasses used in this work use a minimum pre-treatment, and after drying milling, sieving, and wet milling, they are transformed into a brown and homogeneous suspension of fine particles with a broad distribution of size (mostly particles up to 5 μm in diameter) [30]. Thus, we believe that the small size of these particles may facilitate the action of cellulases. However, the low hydrolysis capacity of the enzymes on sugarcane bagasse may have been caused by a process of adsorption to the lignin present. To have a better understanding of this phenomenon, a more detailed characterization of the composition of the various sources used for hydrolysis and energy production would be necessary. These biomasses have different lignin compositions and non-specific binding of cellulases to lignin has been implicated as the main factor in the loss of cellulose activity during biomass conversion to sugars.
Thus, depending on the composition of the biomass and the specificity of the enzyme, these can interact in different ways, which leads to differences in hydrolysis ability. Specific enzyme adsorption to lignin from a mixture of biomass hydrolyzing enzymes is a competitive affinity and both hydrophobic and electrostatic interactions are responsible for this binding phenomenon [56] and lignin from different plant origins coupled with various pre-treatment chemistries might result in a variable adsorption surface chemistry and enzyme accessibility [57].
We demonstrated that PcβGlu2 is more efficient than Pcβglu1 in this kind of application. Pcβglu2 was able to hydrolyze substrates tested without the addition of any other enzyme and also supplemented T. reesei cocktail, increasing the final hydrolysis percentage of banana pseudostem. It is important to notice that, from the three biomass sources used, banana pseudostem has the lower content of lignin, around 9% [58], in comparison with sugarcane (17–24%) [59] and coconut (up to 42%) [60]. In this way, the observation of a better performance of Pcβglu2 when supplementing T. reesei enzymes in the hydrolysis of banana pseudostem, in comparison with the other sources of biomass tested, might be related to a lower lignin content. Surprisingly, P. citrinum enzymes showed a significant activity against coconut fiber, the biomass with the highest content of lignin among the tested, and when Pcβglu2 was used together with T. reesei cocktail, the final quantity of reducing sugars released from this material increased almost twice. Nevertheless, these data must be considered with caution, because the results in some cases suggest inhibition when the different enzymes sources are mixed (e.g., sugarcane bagasse in Fig. 9a, d) or an insignificant activation at the initial times of hydrolysis (coconut fiber in Fig. 9c, f).
As a whole, all these characteristics indicate that the enzymes secreted by P. citrinum, especially Pcβglu2, might be reliable candidates to improve the hydrolysis of specific biomass sources. As noted by kcat values and subsite affinity mapping, these enzymes were capable of hydrolyzing a broad range of oligosaccharides with high catalytic efficiency. The concerted action of exo- and endo-cellulases, together with hemicellulases, might result in temporary accumulation of oligosaccharides with intermediate size, or even disaccharides as laminaribiose, which might inhibit cellulases. In this respect, as far as biorefinery process is concerned, it is interesting that the β-glucosidases used possess also specificity for di/oligosaccharides other than cellobiose because of the use of heterogeneous biomass feedstocks.
Conclusions
Penicillium citrinum UFV1 produces two β-glucosidases when cultivated under SMF using wheat bran as a carbon source. These enzymes use a double displacement hydrolysis mechanism and might belong to GH3 family. The purified enzymes, PcβGlu1 and PcβGlu2, have different molecular and biochemical characteristics, at least five subsites for glycosyl residue binding in their active site, and have a broad substrate specificity. Both enzymes showed relevant features to be applied in biotechnological processes, such as biomass saccharification. PcβGlu2 showed an interesting potential by acting synergically with T. reesei cocktail in the hydrolysis of very complex and lignin-rich biomass, as colloidal banana pseudostem and coconut fiber.
Methods
Reagents
The reagents p-nitrophenyl β-d-glucopyranoside (ρNPβGlu), methylumbelliferyl-β-glucopyranoside (MUβGlu), octyl-β-glucopyranoside (octylβGlu), gentiobiose, laminaribiose, cellobiose, cellotriose, cellotetraose, cellopentaose, microcrystalline cellulose (Avicel®, Cat 11365), low viscosity carboxymethylcellulose (CMC, Cat C5678), ρ-nitrophenyl-β-d-galactopyranoside (ρNPβgal), methylumbeliferil-β-d-mannopyranoside (MUβMan), ρ-nitrophenol, 4-methylumbelliferone, cellulases from T. reesei ATCC 26921 (Cat. C8546) and the molecular markers Blue dextran (Cat. D4772), Cytochrome C (Cat. 12 kDa), carbonic anhydrase (Cat. C7025), serum albumin bovine (Cat. A8531), alcohol dehydrogenase (Cat. A8656), and beta amylase (Cat. A8781) were purchased from Sigma-Aldrich Company (St. Louis, Missouri, USA). Trypsin (Cat. V511A) for mass spectrometry was acquired from Promega Corporation (Madison, Wisconsin, USA). Glucose oxidase-based glucose quantitation reagent was acquired from Bioclin (Minas Gerais, Brazil). Other reagents used in this work were analytical grade.
Organism and culture conditions
Penicillium citrinum UFV1 was obtained from the mycological collection of the Seed Pathology and Post Harvest Laboratory, Federal University of Viçosa, MG, Brazil. The fungus was maintained on potato dextrose agar (PDA) at 4 °C. To evaluate enzyme induction, fungus was maintained in PDA plates for 7 days at 28 °C for sporulation, and spores from this culture were inoculated into 1000 mL Erlenmeyer flasks containing 400 mL of minimal requirement (MR) medium at a concentration of 107 spores mL−1 after sterilization. The MR medium consisted of 0.3% K2HPO4, 1.05% KH2PO4, 0.015% MgSO4·7H2O, 0.15% (w/v) (NH4)2SO4, 0.09% yeast extract, and 1% of wheat bran as carbon source [61]. The culture was incubated at 28 °C and 180 rpm for 240 h. The soluble fraction was separated by filtration through nylon cloth and centrifugation at 21,000×g for 15 min at 4 °C and used as a source of secreted enzymes.
Enzymatic assays and protein quantitation
Unless otherwise specified, β-glucosidase activities were assayed at 40 °C in 50 mM sodium acetate buffer, pH 5.0. Assays with substrates 0.5 mM p-nitrophenyl β-d-glucopyranoside (pNPβGlu), 0.5 mM p-nitrophenyl β-d-galactopyranoside (pNPβGal), 14 µM methylumbelliferyl-β-glucopyranoside (MUβGlu), or 14 µM methylumbelliferyl-β-mannopyranoside (MuβMan) were interrupted after different time intervals by adding 0.25 M Na2CO3. The amount of p-nitrophenol (pNP) released was determined at 410 nm and the amount of 4-methylumbelliferone released was determined at 355 nm excitation and 460 nm emission. Assays with 0.5 mM cellobiose, cellotriose, cellotetraose, cellopentaose, laminaribiose, gentiobiose, or octyl-β-glycopyranoside were interrupted at different time intervals by incubating the mixture at 99 °C for 5 min. The amount of glucose released was determined at 505 nm according to glucose oxidase method [62]. Assays against 0.25% laminarin, xylan, microcrystalline cellulose (Avicel®), or carboxymethylcellulose (CMC) were interrupted after different time intervals by incubating the mixture at 99 °C for 5 min in a thermocycler (Applied Biosystems, Singapore). Reducing sugars released were determined as previously described [63, 64]. Controls without enzyme or substrate were included. One unit of enzymatic activity (U) was defined as the amount of enzyme which released 1 μmol of product/min.
Protein concentrations were determined with the BCA method [65]; for samples containing ammonium sulfate, they were treated with deoxycholic acid previously to BCA assay [66]. For purified samples, protein concentration was determined according to the principle of silver binding [67].
Purification of β-glucosidases
Hydrophobic interaction chromatography
Solid (NH4)2SO4 was added to the soluble fraction of P. citrinum liquid cultures (see “Organism and culture conditions” section) to achieve 50% saturation. After 30 min at 4 °C with mixing, sample was submitted to centrifugation at 21,000 g for 15 min at 4 °C; the precipitate was discarded and 5 mL of supernatant was applied to a HiTrap Phenyl FF column (2.5 cm × 1.6 cm) (AKTA Prime Plus, GE, Uppsala, Sweden) equilibrated in 50 mM sodium acetate pH 5 containing (NH4)2SO4 2.2 M (Buffer A). Elution was undertaken with 2.2–0 M (NH4)2SO4 gradient by mixing Buffer A with increasing amounts of 50 mM sodium acetate pH 5, at 2 mL min−1. Fractions of 5 mL each were collected and assayed using MUβGlu and pNPβGlu substrates. The more active fractions (12–16, named PcβGlu1 and 25–30, named PcβGlu2) were pooled and stored at − 20 °C for further analysis.
Gel filtration chromatography
Pooled fractions corresponding to PcβGlu2 in the “Hydrophobic interaction chromatography” were applied (0.5 mL) onto a Superdex 200 10/300 GL (1.0 cm × 30 cm) (AKTA Purifier, GE, Uppsala, Sweden) equilibrated in 50 mM sodium acetate pH 5 containing 100 mM (NH4)2SO4 and eluted with the same buffer at 0.5 mL min−1. Fractions of 0.5 mL were collected and assayed using MUβGlu substrate. The more active fractions (29–32) were pooled, BSA 1 mg mL−1 was added for stabilization and this material was stored at 4 °C until analysis.
SDS-PAGE, PAGE, and relative molecular mass measurements
Samples were precipitated with TCA [68] and SDS-PAGE (7.5%, w/v) analysis of enzyme fractions was performed according to the method of Laemmli and gels were stained with silver nitrate [69, 70].
For nondenaturing polyacrylamide gel electrophoresis (PAGE), samples were desalted by dialysis against 10 mM sodium acetate buffer pH 5.0 and concentrated in Centricon with 30 kDa cutoff (Millipore Corporation, Massachusetts, USA). β-Glucosidase activity was detected on the polyacrylamide gel with MUβGlu 4 mM in sodium acetate buffer [71]. Methylumbelliferone released from MUβGlu was visualized under ultraviolet light.
The apparent native molecular mass of the purified β-glucosidases was estimated using a Superdex 200 10/300 GL (1.0 cm × 30 cm) (GE, Uppsala, Sweden). The column was equilibrated and eluted with 50 mM sodium acetate pH 5 at 0.5 mL min−1. The void volume was determined using Blue dextran (2000 kDa). Cytochrome C (12 kDa), carbonic anhydrase (29 kDa), serum albumin bovine (66 kDa), alcohol dehydrogenase (150 kDa), and beta amylase (200 kDa) were used as molecular mass standards.
Protein identification by mass spectrometry
PcβGlu1 and PcβGlu2 samples were dialyzed against 10 mM sodium acetate buffer pH 5 and concentrated in Concentrator 5301 (Eppendorf, Hamburg, Germany). Desalted enzymes were reduced, alkylated, and subjected to trypsin digestion for 24 h at 37 °C, 1:50 enzyme:substrate (m:m) [72]. Tryptic peptides were desalted with ZipTip C18 (Millipore Corporation, Massachusetts, USA) according to manufacturer instructions. Samples were dried in Concentrator 5301 (Eppendorf, Hamburg, Germany), ressuspended in formic acid 1%, and submitted to MS/MS analysis using an EASY-nLC coupled to a nanoESI LTQ Orbitrap XL instrument (Thermo Scientific, USA). Chromatographic and mass spectrometric analyses’ conditions were the same as previously described [73], although the separation column was only 12-cm long and gradient conditions were: 2–60% B during 52 min; up to 80% B in 4 min, maintaining at this concentration for 2 min more. All MS/MS spectra were analyzed using PEAKS Studio 8.5 build 20180105 (Bioinformatics Solutions, Canada). After data refinement with the precursor (mass only) correction option, PEAKS DE NOVO analysis was run assuming trypsin digestion, with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 15 ppm. Cysteine (C) carbamidomethylation (+ 57.02 Da) was set as fixed modification and the following variable modifications were searched: deamidation at N/Q (+ 0.98 Da), oxidation at M and unspecific carbamidomethylation (+ 57.02) at D, E, H, K, and free N-terminus; a maximum of 2 variable modifications per peptide was allowed. PEAKS DB analysis was performed using these same parameters plus the possibility of up to two missed enzyme cleavages and non-specific cleavage at one side of the peptides. Searches were made against a subdatabase, composed of all Uniprot entries that matched the term “Penicillium” (338,849 entries, downloaded on April 18, 2018). Finally, 310 other possible modifications were searched with the PEAKS PTM algorithm, using the same parameters described above, against a protein subdatabase composed only by protein entries found by the joint PEAKS DE NOVO and PEAKS DB searches. False discovery rates (FDR) were estimated through the PEAKS decoy fusion approach. A peptide-spectrum match FDR of 0.1% and protein identifications with at least 2 unique peptides were the criteria used to establish FDR values at peptide and protein levels ≤ 1%.
Configuration of the anomeric carbon atom of reaction products
To identify the anomeric configuration of the d-glucose released by β-glucosidases [74], enzymes were incubated at 40 °C in 50 mM sodium acetate buffer pH 5 with cellobiose as a substrate. After 30 min for PcβGlu1 and 120 min for PcβGlu2, reactions were stopped by the addition of 100 mM glucono-δ-lactone to the incubation medium, and aliquots of 0.1 mL were removed. Each sample’s aliquot was incubated at 100 °C for 5 min followed by addition of 0.1 mL of glucose oxidase reagent (Bioclin, Minas Gerais, Brazil). The absorbance was recorded continuously at 505 nm for 20 min. Controls were prepared similar to the experimental samples and the substrate was replaced by a fresh solution of α-d-glucose or by a solution of α-d-glucose incubated at 100 °C for 5 min.
Kinetic studies
The effect of pH on enzyme stability and activity was determined at 40 °C using the following buffers (100 mM): for PcβGlu1, citrate/phosphate (pH 3–7), EPPS (pH 7–9), AMPSO (pH 9–10), CAPS (pH 10–11) and for PcβGlu2, sodium acetate (pH 3.7–6), MOPS (pH 6–8), EPPS (pH 7–9), and AMPSO (pH 9–10). For pH stability assays, enzymes were pre-incubated in the determined pH for 4 h at 40 °C to residual activity determination according to “Enzymatic Assays and Protein Quantitation” section MUβGlu as a substrate.
Thermostability was evaluated by incubating PcβGlu1 in the presence of ammonium sulfate and PcβGlu2 in the presence of BSA 1 mg mL−1 for 4 h at temperatures from 30 to 80 °C. PcβGlu1 was also incubated at 50 °C in the absence of ammonium sulfate and PcβGlu2 incubated at 50 °C in the absence of BSA. The residual activities were determined according to “Enzymatic Assays and Protein Quantitation” section using MUβGlu as a substrate. Half-life (t1/2) and inactivation constant (kd) were calculated from a linear plot of log (residual activity) versus incubation time [75].
Effect of metal ions
The effect of NaCl, NaNO3, KCl, NH4Cl, Pb(NO3)2, BaCl2, ZnCl2, MgCl2, MnCl2, CaCl2, CoCl2, FeCl3, HgCl, and CuSO4 on PcβGlu1 and PcβGlu2 activity was determined by pre-incubating enzymes for 30 min at 40 °C in 50 mM sodium acetate buffer pH 5 in the presence of 100 mM of each salt above. Residual activities were determined as described above using MUβGlu as a substrate.
Determination of kinetic parameters
The values of the Michaelis constant (Km) and the maximum velocity (Vmax) were determined for PcβGlu1 and PcβGlu2 incubating enzymes in 50 mM sodium acetate buffer pH 5.0 at 40 °C with pNPβGlu, MUβGlu, OctylβGlu, gentiobiose, laminaribiose, cellobiose, cellotriose, cellotetraose, cellopentaose, or laminarin at concentrations ranging from 0.04 to 20 mM depending on substrate. Inhibition of PcβGlu1 and PcβGlu2 by glucose (0–15 mM) and cellobiose (0–10 mM) was determined in the presence of pNPβGlu as the substrate. Activity was determined depending on the substrate as described above. Values for Km, Vmax, and Ki were determined using the GraFit software (GraFit version_7.0.3, Erithacus Software Limited) and Michaelis–Menten equation [75]. When high substrate concentration inhibition was detected, kinetic parameters were calculated taking into account the reaction below (Scheme 1) [76]:
Subsite mapping
The subsite affinities of β-glucosidases were evaluated according to the Hiromi’s method [29]. Subsite affinities Ai (kcal mol−1) for glucose and kint (catalytic rate independent of the polymerization degree of the substrate) were calculated from Km values and catalytic rate constants (kcat) for the hydrolysis of cellodextrins with polymerization degrees from 2 to 5. The validity of the calculations was subsequently confirmed by comparing experimental and theoretical kcat, Km, and kcat/Km values following Hiromi’s method to estimate the validity of Ai and kint values obtained with errors of less than 20%. A2 was calculated by the method II. The theoretical values were obtained according to the equations:
where n is the substrate degree of polymerization, 0.021 corresponds to the contribution of the mixing entropy in water at 40 °C [29], R is the gas constant, and T is the absolute temperature.
Biomass saccharification
Penicillium citrinum UFV1 purified β-glucosidases (PcβGlu1 and PcβGlu2) and commercial cellulase from T. reesei ATCC 26921 were applied in saccharification experiments using different sources of biomass. Colloidal suspensions of sugar cane bagasse, coconut fiber, and banana pseudostem were prepared with a minimum pre-treatment [30]. PcβGlu1 and PcβGlu2 were concentrated in Centricon 30 (Millipore, Billerica, MA, USA) membrane filter. Enzymatic saccharification of colloidal biomasses was performed at an initial solid concentration of 2.5% (w/v) in 50 mM sodium acetate buffer at pH 5. Enzyme loading was 10 CBU/g biomass of PcβGlu1 or PcβGlu2 and 7 CBU/g biomass for T. reesei cellulase. Sodium azide 0.01% (w/v) was added to the reaction mixture to inhibit microbial growth. The reaction was carried out in an orbital shaker at 150 rpm and 40 °C for 24 h. Samples (0.5 mL) were taken from the reaction mixture at different time intervals, immediately heated to 100 °C to denature enzymes, cooled, and then centrifuged for 5 min at 21,000 g. The released reducing sugars in supernatant and total sugar at colloidal biomass were detected by the previous described methods [77, 78]. The saccharification hydrolysis rate was estimated comparing the reducing sugar released at saccharification with the quantity of total sugars at colloidal biomass. Statistical analysis graphs and data analysis were performed with the software GraphPad Prism version 5.01 for Windows. Two-way ANOVA was used followed by Tukey’s multiple comparison tests, p < 0.05.
Abbreviations
- PcβGlu1:
-
Penicillium citrinum UFV1 β-glucosidase 1
- PcβGlu2:
-
Penicillium citrinum UFV1 β-glucosidase 2
- pNPβGlu:
-
p-nitrophenyl β-d-glucopyranoside
- pNPβGal:
-
p-nitrophenyl β-d-galactopyranoside
- MUβGlu:
-
methylumbelliferyl-β-glucopyranoside
- MuβMan:
-
methylumbelliferyl-β-manopyranoside
- OctylβGlu:
-
octyl-β-glucopyranoside
- CMC:
-
carboxymethylcellulose
- EPPS:
-
4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- AMPSO:
-
N-(1,1-dimethyl-2-hydroxyethyl)-3-amino-2-hydroxypropanesulfonic acid
- CAPS:
-
3-(cyclohexylamino)-1-propanesulfonic acid
- MOPS:
-
4 morpholinepropanesulfonic
- TCA:
-
trichloroacetic acid
- BCA:
-
bicinchoninic acid
- BSA:
-
Bovine Serum Albumin
References
Singhania RE, Patel A, Sukumaran RK, Larroche C, Pandey A. Role and significance of beta-glucosidases in the hydrolysis of cellulose for bioethanol production. Bioresour Technol. 2013;127:500–7.
Harnpicharnchai P, Champreda V, Sornlake W, Eurwilaichitr L. A thermotolerant β-glucosidase isolated from an endophytic fungi, Periconia sp., with a possible use for biomass conversion to sugars. Protein Express Purif. 2009;67:61–9.
Bhatia Y, Mishra S, Bisaria VS. Microbial β-glucosidases: cloning, properties and applications. Crit Rev Biotechnol. 2002;22:375–407.
Ng I, Li C, Chan S, Chir J, Chen PT, Tong C, Yu S, Ho TD. High-level production of a thermoacidophilic β-glucosidase from Penicillium citrinum YS40-5 by solid-state fermentation with rice bran. Bioresour Technol. 2010;10:1310–7.
Pal S, Banik SP, Ghorai S, Chowdhury S, Khowala S. Purification and characterization of a thermostable intra-cellular β-glucosidase with transglycosylation properties from filamentous fungus Termitomyces clypeatus. Bioresour Technol. 2010;101:2412–20.
Sawant S, Birhade S, Anil A, Gilbert H, Lali A. Two-way dynamics in β-glucosidase catalysis. J Mol Catal B Enzymatic. 2016;133:161–6.
Bhat MK, Bhat S. Cellulose degrading enzymes and their potential industrial applications. Biotechnol Adv. 1997;15:583–620.
Ducret A, Trani M, Lortie R. Screening of various glycosidases for the synthesis of octyl glucoside. Biotechnol Bioeng. 2002;77:752–7.
Gueguen Y, Chemardin P, Labrot P, Arnaud A, Galzy P. Purification and characterization of an intracellular β-glucosidase from a new strain of Leuconostoc mesenteroides isolated from cassava. J Appl Microbiol. 1997;82:469–76.
Tokpohozin SE, Fischer S, Sacher B, Becker T. β-d-Glucosidase as “key enzyme” for sorghum cyanogenic glucoside (dhurrin) removal and beer bioflavouring. Food Chem Toxicol. 2016;97:217–23.
Steensels J, Daenen L, Malcorps P, Derdelinckx G, Verachtert H, Verstrepen KJ. Brettanomyces yeasts—from spoilage organisms to valuable contributors to industrial fermentations. Int J Food Microbiol. 2015;206:24–38.
Çelik A, Dinçer A, Aydemir T. Characterization of β-glucosidase immobilized on chitosan-multi walled carbon nanotubes (MWCNTS) and their application on tea extracts for aroma enhancement. Int J Biol Macromol. 2016;89:406–14.
Bayer EA, Lamed R, Himmel ME. The potential of cellulases and cellulosomes for cellulosic waste management. Curr Opin Biotechnol. 2007;18:237–45.
Limayem A, Ricke SC. Lignocellulosic biomass for bioethanol production: current perspectives, potential issues and future prospects. Prog Energy Combust Sci. 2012;38:449–67.
Cheng KK, Cai BY, Zhang JA, Ling HZ, Zhou YJ, Ge JP, et al. Sugarcane bagasse hemicelluloses hydrolysate for ethanol production by acid recovery process. Biochem Eng J. 2008;38:105–9.
Kim S, Dale BE. Global potential bioethanol production from wasted crops and crops residues. Biomass Bioenerg. 2005;29:361–75.
Knauf M, Moniruzzaman M. Lignocellulosic biomass processing. Persp Int Sugar J. 2004;106:147–50.
Kadam KL, McMillan JD. Availability of corn stover as a sustainable feedstock for bioethanol production. Bioresour Technol. 2003;88:17–25.
Ng I, Tsai S, Ju Y, Yu S, Ho TD. Dynamic synergistic effect on Trichoderma reesei cellulases by novel β-glucosidases from Taiwanese fungi. Bioresour Technol. 2011;102:6073–81.
Xia W, Xu X, Qian L, et al. Engineering a highly active thermophilic β-glucosidase to enhance its pH stability and saccharification performance. Biotechnol Biofuels. 2016;9:147.
Yang S, Jiang Z, Yan Q, Zhu H. Characterization of a thermostable extracellular β-glucosidase with activities of exoglucanase and transglycosylation from Paecilomyces thermophila. J Agric Food Chem. 2008;56:602–8.
Andrade LGA, Maitan-Alfenas GP, Gomes KS, Falkoski DL, Alfenas RF, Guimarães VM. Sugarcane bagasse saccharification by purified β-glucosidases from Chrysoporthe cubensis. Biocatal Agric Biotechnol. 2017;12:199–205.
Teugjas H, Väljamäe P. Selecting β-glucosidases to support cellulases in cellulose saccharification. Biotechnol Biofuels. 2013;6:105.
Akiyama T, Shibuya N, Hrmova M, Fincher GB. Purification and characterization of a (1– > 3)-beta-d-glucan endohydrolase from rice (Oryza sativa) bran. Carbohydr Res. 1997;297:365–74.
Saha SP, Ghosh S. Optimization of xylanase production by Penicillium citrinum xym2 and application in saccharification of agro-residues. Biocatal Agric Biotechnol. 2014;3:188–96.
Dutta T, Sahoo R, Sengupta R, Ray SS, Bhattacharjee A, Ghosh S. Novel cellulases from an extremophilic filamentous fungi Penicillium citrinum: production and characterization. J Ind Microbiol Biotechnol. 2008;35:275–82.
Arakawa T, Timasheff SN. Mechanism of protein salting in and salting out by divalent cation salts: balance between hydration and salt binding? Biochemistry. 1984;23:5912–23.
Cobos A, Estrada P. Effect of polyhydroxylic cosolvents on the thermostability and activity of xylanase from Trichoderma reesei QM 9414. Enzyme Microb Technol. 2003;33:810–8.
Hiromi K, Nitta Y, Numata C, Ono S. Subsite affinities of glucoamylase: examination of the validity of the subsite theory. Biochim Biophys Acta. 1973;302:362–75.
Lucena SA, Lima SL, Cordeiro LSA Jr, Sant’Anna C, Constantino R, Azambuja P, de Souza W, Garcia ES, Genta FA. High throughput screening of hydrolytic enzymes, from termites using a natural substrate derived from sugarcane bagasse. Biotechnol Biofuels. 2011;4:51.
Wang L, Ridgway D, Gu T, Moo-Young M. Bioprocessing strategies to improve heterologous protein production in filamentous fungal fermentations. Biotechnol Adv. 2005;23:115–29.
Falkoski DL, Guimarães VM, de Almeida MN, Alfenas AC, Colodette JL, de Rezende ST. Chrysoporthe cubensis: a new source of cellulases and hemicellulases to application in biomass saccharification processes. Bioresour Technol. 2013;130:296–305.
Brijwani K, Oberoi HS, Vadlani PV. Production of a cellulolytic enzyme system in mixed-culture solid-state fermentation of soybean hulls supplemented with wheat bran. Process Biochem. 2010;45:120–8.
Jeya M, Joo AR, Lee KM, Tiwari MK, Lee KM, Kim SH, Lee JK. Characterization of beta-glucosidase from a strain of Penicillium purpurogenum KJS506. Appl Microbiol Biotechnol. 2010;86:1473–84.
Liu J, Yuan X, Zeng G, Shi J, Chen S. Effect of biosurfactant on cellulose and xylanase production by Trichoderma viride in solid substrate fermentation. Process Biochem. 2006;41:2347–51.
Camassola M, Dillon AJP. Production of cellulases and hemicellulases by Penicillium echinulatum grown on pretreated sugar cane bagasse and wheat bran in solid-state fermentation. J Appl Microbiol. 2007;103:2196–204.
Souza FH, Nascimento CV, Rosa JC, Masui DC, Leone FA, Jorge JA, Furriel RPM. Purification and biochemical characterization of a mycelial glucose- and xylose-stimulated β-glucosidase from the thermophilic fungus Humicola insolens. Process Biochem. 2010;45:272–8.
Christakopoulos P, Kekos D, Marcris BJ, Goodenough PW, Bhat MK. Purification and characterization of an extracellular β-glucosidase with transglycosylation and exo-glucosidase activities from Fusarium oxysporum. Biotech Lett. 1994;16:587–92.
Collins T, Gerday C, Feller G. Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev. 2005;29:3–23.
Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:233–8.
Varghese JN, Garrett TPJ, Colman PM, Chent L, Hoj PB, Fincher GB. Three-dimensional structures of two plant b-glucan endohydrolases with distinct substrate specificities. Proc Nati Acad Sci USA. 1994;91:2785–9.
Pozzo T, Pasten JL, Karlsson EN, Logan DT. Structural and functional analyses of β-glucosidase 3B from Thermotoga neapolitana: a thermostable three-domain representative of glycoside hydrolase 3. J Mol Biol. 2010;397:724–39.
Yoshida E, Hidaka M, Fushinobu S, Koyanagi T, Minami H, Tamaki H, Kitaoka M, Katayama T, Kumagai H. Role of a PA14 domain in determining substrate specificity of a glycoside hydrolase family 3 β-glucosidase from Kluyveromyces marxianus. Biochem J. 2010;431:39–49.
Varrot A, Schulein M, Davies GJ. Insights into ligand-induced conformational change in Cel5A from Bacillus agaradhaerens revealed by a catalytically active crystal form. J Mol Biol. 2000;297:819–28.
Korotkova OG, Semenova MV, Morozova VV, Zorov IN, Sokolova LM, Bubnova TM, Okunev ON, Sinitsyn AP. Isolation and properties of fungal beta-glucosidases. Biochem. 2009;74:569–77.
Park A, Hong JH, Kim J, Yoon J. Biochemical characterization of an extracellular β-glucosidase from the fungus, Penicillium italicum, isolated from rotten citrus peel. Mycobiology. 2012;40:173–80.
Karnchanatat A, Petsom A, Sangvanich P, Piaphukiew J, Whalley AJ, Reynolds CD, Sihanonth P. Purification and biochemical characterization of an extracellular beta-glucosidase from the wood-decaying fungus Daldinia eschscholzii (Ehrenb.:Fr.) Rehm. FEMS Microbiol Lett. 2007;270:162–70.
Hrmova M, De Gori R, Smith BJ, Fairweather JK, Driguez H, Varghese JN, Fincher GB. Structural basis for broad substrate specificity in higher plant beta-d-glucan glucohydrolases. Plant Cell. 2002;14:1033–52.
Langston J, Sheehy N, Xu F. Substrate specificity of Aspergillus oryzae family 3 β-glucosidase. Biochim Biophys Acta Proteom. 2006;1764:972–8.
Chauve M, Mathis H, Huc D, Casanave D, Monot F, Ferreira NL. Comparative kinetic analysis of two fungal β-glucosidases. Biotechnol Biofuels. 2010;3:3–10.
Bohlin C, Praestgaard E, Baumann MJ, Borch K, Praestgaard J, Monrad RN, Westh P. A comparative study of hydrolysis and transglycosylation activities of fungal β-glucosidases. Appl Microbiol Biotechnol. 2013;97:159–69.
Seidle HF, Allison SJ, George E, Huber RE. Trp-49 of the family 3 beta glucosidase from Aspergillus niger is important for its transglucosidic activity: creation of novel beta-glucosidases with low transglucosidic efficiencies. Arch Biochem Biophys. 2006;455:110–8.
Thongpoo P, McKee LS, Araújo AC, Kongsaeree PT, Brumer H. Identification of the acid/base catalyst of a glycoside hydrolase family 3 (GH3) β-glucosidase from Aspergillus niger ASKU28. Acta Biochim Biophys. 2013;1830:2739–49.
Vassilev SV, Baxter D, Andersen LK, Vassileva CG. An overview of the chemical composition of biomass. Fuel. 2010;89:913–33.
Werther J, Saenger M, Hartge EU, Ogada T, Siagi Z. Combustion of agricultural residues. Prog Energy Combust Sci. 2000;26:1–27.
Yarbrough JM, Mittal A, Mansfield E, Taylor LE, Hobdey SE, Sammond DW, Bomble YJ, Crowley MF, Decker SR, Himmel ME, Vinzant TB. New perspective on glycoside hydrolase binding to lignin from pretreated corn stover. Biotechnol Biofuels. 2015;8:214.
Nakagame S, Chandra RP, Saddler JN. The effect of isolated lignins, obtained from a range of pretreated lignocellulosic substrates, on enzymatic hydrolysis. Biotechnol Bioeng. 2010;105:871–9.
Gabhane J, William SPMP, Gadhe A, Rath R, Vaidya AN, Wate S. Pretreatment of banana agricultural waste for bio-ethanol production: individual and interactive effects of acid and alkali pretreatments with autoclaving, microwave heating and ultrasonication. Waste Manag. 2014;34:498–503.
Masarin F, Gurpilhares DB, Baffa DCF, Barbosa MHP, Carvalho W, Ferraz A, Milagres AMF. Chemical composition and enzymatic digestibility of sugarcane clones selected for varied lignin content. Biotechnol Biofuels. 2011;4:55.
Muensri P, Kunanopparat T, Menut P, Siriwattanayotin S. Effect of lignin removal on the properties of coconut coir fiber/wheat gluten biocomposite. Compos Part A Appl Sci Manuf. 2011;42:173–9.
Mandels M, Weber J. The production of cellulases. Adv Chem. 1969;95:391–414.
Raabo E, Terkildsen TC. On the enzymatic determination of blood glucose. Scand J Clin Lab Invest. 1960;12:402–7.
Fox JD, Robyt JF. Miniaturization of three carbohydrate analyses using a microsample plate reader. Anal Biochem. 1991;195:93–6.
Lucena SA, Moraes CS, Costa SG, de Souza W, Azambuja P, Garcia ES, Genta FA. Miniaturization of hydrolase assays in thermocyclers. Anal Biochem. 2013;434:39–43.
Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–86.
Rhoderick E, Brown KL, Jarvis KJ, Hyland KJ. Protein measurement using bicinchoninic acid: elimination of interfering substances. Anal Biochem. 1989;180:136–9.
Krystal G, Macdonald C, Munt B, Ashwell S. A method for quantitating nanogram amounts of soluble protein using the principle of silver binding. Anal Biochem. 1985;148:451–60.
Yvon M, Chabanet C, Pélissier JP. Solubility of peptides in trichloroacetic acid (TCA) solutions. Int J Pept Protein Res. 1989;34:166–76.
Laemmli UK. Cleavage of structural proteins during the assembly of head of bacteriophage T4. Nature. 1970;227:680–3.
Morrissey JH. Silver stain for proteins in polyacrylamide gel. A modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981;11:307–10.
Badhan AK, Chadha BS, Kaur J, Saini HS, Bhat MK. Production of multiple xylanolytic and cellulolytic enzymes by thermophilic fungus Myceliophthora sp. IMI 387099. Bioresour Technol. 2007;98:504–10.
Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–906.
Nicolau CA, Carvalho PC, Junqueira-de-Azevedo IL, Teixeira-Ferreira A, Junqueira M, Perales J, Neves-Ferreira AG, Valente RH. An in-depth snake venom proteopeptidome characterization: benchmarking Bothrops jararaca. J Proteomics. 2017;151:214–31.
Semenza G, Curtis HC, Raunhardt O, More P, Muler M. The configurations at the anomeric carbon of the reaction products of some digestive enzymes. Carbohyd Res. 1969;10:417–28.
Segel IH. Enzyme kinetics. Behavior and analysis of rapid equilibrium and steady-state enzyme systems. New York: Willey; 1975.
Cleland WW. Substrate inhibition. Methods Enzymol. 1979;63:500–13.
Miller GL. Use of dinitrosalicyclic acid reagent for determination of reducing sugars. Anal Chem. 1959;31:426–8.
Dubois M, Gilles KA, Hamilton JK, Rebers PA, Smith F. Colorimetric method for determination of sugars and related substances. Anal Chem. 1956;28:350–6.
Authors’ contributions
SGC carried out the experiments, analyzed the data, and drafted the manuscript. OLP, AT-F, and RHV designed experiments and carried out experiments. STR, VMG, RHV, and FAG conceived, supervised the study, and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was funded by the Brazilian research agencies Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Fundação de Amparo a Pesquisa do Estado de Minas Gerais (FAPEMIG), Fundação Oswaldo Cruz (FIOCRUZ), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Table S1.
Production of cellulases and hemicellulases by Penicillium citrinum cultivated in liquid media using wheat bran as carbon source. Activities are presented as U/mL ± SD. CMC—carboxymethylcellulose. Superscript numbers represent the number of days in culture to achieve the maximum production of each enzyme.
Additional file 2: Table S2.
Effect of solutes on PcβGlu2 stability.
Additional file 3: Table S3.
Summary of protein groups identified following mass spectrometry analysis of purified PcBglu1. GH—match to Glycoside Hydrolase Family (E.C.3.2.1.21).
Additional file 4: Table S4.
Summary of protein groups identified following mass spectrometry analysis of partially purified PcBglu2. GH—match to Glycoside Hydrolase Family (E.C.3.2.1.21).
Additional file 5: Table S5.
Comparison between experimental and theoric rate parameters for PcβGlu1 and PcβGlu2 of P. citrinum.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
da Costa, S.G., Pereira, O.L., Teixeira-Ferreira, A. et al. Penicillium citrinum UFV1 β-glucosidases: purification, characterization, and application for biomass saccharification. Biotechnol Biofuels 11, 226 (2018). https://doi.org/10.1186/s13068-018-1226-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-018-1226-5