Abstract
Background
Triacylglycerols (TAGs) rich in medium-chain fatty acids (MCFAs, C10–14 fatty acids) are valuable feedstocks for biofuels and chemicals. Natural sources of TAGs rich in MCFAs are restricted to a limited number of plant species, which are unsuitable for mass agronomic production. Instead, the modification of seed or non-seed tissue oils to increase MCFA content has been investigated. In addition, microbial oils are considered as promising sustainable feedstocks for providing TAGs, although little has been done to tailor the fatty acids in microbial TAGs.
Results
Here, we first assessed various wax synthase/acyl-coenzyme A:diacylglycerol acyltransferases, phosphatidic acid phosphatases, acyl-CoA synthetases as well as putative fatty acid metabolism regulators for producing high levels of TAGs in Escherichia coli. Activation of endogenous free fatty acids with tailored chain length via overexpression of the castor thioesterase RcFatB and the subsequent incorporation of such fatty acids into glycerol backbones shifted the TAG profile in the desired way. Metabolic and nutrient optimization of the engineered bacterial cells resulted in greatly elevated TAG levels (399.4 mg/L) with 43.8% MCFAs, representing the highest TAG levels in E. coli under shake flask conditions. Engineered cells were observed to contain membrane-bound yet robust lipid droplets.
Conclusions
We introduced a complete Kennedy pathway into non-oleaginous E. coli towards developing a bacterial platform for the sustainable production of TAGs rich in MCFAs. Strategies reported here illustrate the possibility of prokaryotic cell factories for the efficient production of TAGs rich in MCFAs.
Similar content being viewed by others
Background
Worldwide, demand for oil-based products used in the energy industry has increased over recent decades leading to concerns for both the supply and the carbon footprint on the environment. Renewable plant oils, non-seed oils, and microbial oils (or single-cell oils, SCOs) are expected to play vital roles in providing a sustainable source of oils [1, 2]. Triacylglycerols (TAGs) rich in medium-chain fatty acids (MCFAs), including capric acid (C10:0), lauric acid (C12:0), and myristic acid (C14:0), are of particular interest due to their lower freezing point and higher carbon conversion yield. Such oils are important constituents of certain oleochemicals and jet fuels [3,4,5]. However, the end products of wild-type plant and bacterial fatty acid synthetase activities are mainly C16 and C18 fatty acids. Meanwhile, natural sources of oils rich in MCFAs are restricted to a limited number of plant species, such as Cinnamomum camphora, Cuphea sp., Cocos nucifera, and Umbellularia california, which are unsuitable for mass agronomic production [6].
Several studies have investigated the modification of seed oils to increase MCFA content. Co-expression of MCFA-specific CpuDGAT1, CvFatB1, and CvLPAT2 resulted in 25 mol% of C10 fatty acid in Camelina seed oils with modest effects on seed oil content and plant growth [7]. In addition, the accumulation of MCFAs in Nicotiana benthamiana leaf oils has been recently achieved by introduction of MCFA-specific EgDGAT along with CcFatB or UcFatB [8]. Although microbes including Escherichia coli and Yarrowia lipolitica have been engineered to produce medium-chain free fatty acids, fatty alcohols, fatty acid ethyl esters (FAEEs), or fatty alkanes with varying outcomes [9,10,11], the accumulation of MCFAs in the form of TAGs is rare in microorganisms, especially prokaryotic microorganisms.
Before achieving accumulation of TAGs rich in MCFAs, TAGs first needs to be synthesized to a high level. The bacterial Kennedy pathway for TAG biosynthesis involves the sequential esterification of glycerol-3-phosphate to produce a central molecular phosphatidic acid (PA) [12]. PA can be dephosphorylated by phosphatidic acid phosphatase (PAP) to produce diacylglycerol (DAG). DAG is subsequently catalyzed into TAG by wax ester synthase/acyl-coenzyme A:diacylglycerol acyltransferase (WS/DGAT). This enzyme is the rate-limiting enzyme, because it leads to the last step of TAG formation. WS/DGATs are unique in oleaginous microorganisms and a few oil crops [13, 14]. Unlike DGAT1 or DGAT2 from eukaryotic algae, plants, and animals, WS/DGAT is the only class of DGAT responsible for TAG formation in bacteria [15]. Among the reported WS/DGATs, AtfA from Acinetobacter baylyi has been evaluated as the best candidate for TAG production in E. coli [16]. Soon after this report, Atf8 from Rhodococcus jostii RHA1 was demonstrated to efficiently facilitate TAG accumulation [17]. In addition, a thermophilic tDGAT from Thermomonospora curvata was shown to yield over 30% of TAGs to neutral lipids in 30 min in E. coli [18]. It would be interesting to compare the abilities of these newly isolated WS/DGATs to atfA.
In contrast to WS/DGATs, only a few bacterial PAPs have been characterized for TAG synthesis [19]. Heterologous expression of PAP encoding genes from oleaginous Streptomyces coelicolor (lppα or lppβ), or from R. jostii RHA1 (ro00075) has resulted in a two-to-sixfold increase in DAG content in E. coli [20, 21]. Furthermore, the combination of LPPα/β along with WS/DGAT Sco0958 leads to TAG accumulation in E. coli BL21(DE3)∆dgkA∆fadE. Both LPPα and LPPβ are considered to be dedicated PAPs involved in generating DAG for TAG biosynthesis, although LPPβ has a higher PAP activity [21]. The PAP encoding gene pgpB is also present in E. coli, but its activity is generally low [22]; however, PgpB has been used for enhancement of TAG production [16].
Acyl-CoA synthetase, responsible for esterification of free fatty acids (FFAs) to acyl-CoA, is crucial for reconstruction of the acyl-CoA-dependent TAG synthesis pathway in E. coli. There is only a single acyl-CoA synthetase (FadD) with broad substrate specificity identified in E. coli [23]. Overexpression of fadD enhances the TAG content in engineered E. coli [16, 24]. By contrast, numerous acyl-CoA synthetase isoforms are characterized from TAG-producing plants, yeasts, algae, or bacteria [15, 25, 26]. In general, mitochondrial-located acyl-CoA synthetase isoforms activate fatty acids for β-oxidation, while endoplasmic reticulum or plastid-localized isoforms contribute to TAG or PL synthesis in eukaryotic cells. However, whether multiple acyl-CoA synthetases exist and which isoforms of these may be responsible for TAG synthesis in oleaginous bacteria have not been determined yet [15].
The central fatty acid metabolism regulator FadR has been well studied and employed in the previous experiments to increase total fatty acids (TFAs), TAGs, or hydroxy fatty acids in E. coli [16, 27]. However, it has also been reported that FadR does not increase the TAG content in E. coli containing 4.5% TAGs (w/w CDW) [28]. Although FadR has dual roles for both stimulating fatty acid synthesis and inhibiting fatty acid degradation, it represses fadD transcription, and thus impairs recycling of FFA for acyl-CoA-dependent TAG synthesis [29].
Enhancement of MCFA flux is aimed at producing TAGs rich in MCFAs. Since both plants and bacteria use a type II FAS system to synthesize fatty acids, many plant MCFA-specific acyl-ACP thioesterases (TEs) have been successfully applied in E. coli for generating high levels of MCFAs [30, 31]. In our recent work, CnFatB3 from Cocos nucifera, CcFatB1 from Cinnamomum camphora, or CpFatB2 from Cuphea pulcherrima have been shown to greatly enhance the MCFA titer in E. coli BL21(DE3) [32]. In addition, ChFatB2 from Cuphea hookeriana leads to high levels of MCFA accumulation in canola but not in Camelina seeds [33, 34]. Whether ChFatB2 can release MCFAs in E. coli has not been tested yet. In addition, bacterial TE LpFat or CtFat does not enhance the MCFA titer, although the MCFA percentage was indeed increased in our previous experiments [32]. Among several tested bacterial TEs from the oleaginous bacterium A. babylyi, only AcTesA’ led to the accumulation of C14:0 in E. coli, reaching 7.4 µg/mg CDW [35].
In this study, we focused on the metabolic engineering of E. coli aiming to build a sustainable cell platform for the production of TAGs with tailored fatty acids. A Kennedy pathway was reconstructed in E. coli for the accumulation of high levels of the regular TAGs (Fig. 1). MCFA flux was enhanced by a specific TE and redirected into TAGs. Nutritional optimization further improved the production of TAG rich in MCFAs by 63%, reaching its highest level of 399.4 mg/L in shake flasks. In addition, lipid droplets (LDs) with a robust structure were observed in engineered E. coli, suggesting that other value-added lipids may be produced and then stored in the form of prokaryotic LDs with higher stability and less toxicity to cells.
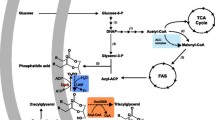
Engineering of E. coli cells for the production of TAG rich in MCFA. Glycolytic pathway, fatty acid biosynthesis II pathway (FASII), fatty acid degradation pathway (β-oxidation), and the reconstructed acyl-CoA-dependent Kennedy pathway are shown in different background colours. Heterologous proteins are highlighted in red. LD lipid droplet, PAP phosphatidic acid phosphatase, MCFAs medium-chain fatty acids, TE acyl-ACP thioesterase, ACS acyl-CoA synthetase, WS/DGAT wax ester synthase/acyl-Coenzyme A:diacylglycerol acyltransferase, AccABCD acetyl-CoA carboxyltransferase, FabA 3-hydroxydecanoyl-ACP dehydrase, FabB 3-ketoacyl-ACP synthase I, FabD malonyl-CoA-ACP transacylase, FabF 3-ketoacyl-ACP synthase II, FabG 3-oxo-acyl-ACP reductase, FabH 3-ketoacyl-ACP synthase III, FabI enoyl-ACP reductase, TesA acyl-CoA thioesterase I, FadA 3-ketoacyl-CoA thiolase, FadB dodecenoyl-CoA delta-isomerase, FadD fatty acyl-CoA synthetase, FadE acyl-CoA dehydrogenase, GlpF glycerol MIP channel, GlpK glycerol kinase, PlsB glycerol-3-phosphate acyltransferase, PlsC 1-acylglycerol-3-phosphate O-acyltransferase, PgpB phosphatidylglycerophosphatase B, DgkA diacylglycerol kinase
Results and discussion
The combination of tDGAT and RoPAP resulted in significant increase inTAGs in E. coli
WS/DGAT and PAP are the two essential enzymes for reconstruction of the Kennedy pathway in E. coli. The expression of AtfA along with RoPAP resulted in an obviously increased TAG, while the expression of AtfA and RjPAP only generated a low amount of TAGs but a higher amount of FAEE and FFA (Fig. 2a). This was consistent with the previous observations that RjPAP was not significantly expressed when TAG accumulated in R. jostii RHA1 [17]. We predicted that RjPAP expression might change the fatty acid composition of the generated DAG, and thus decreased the affinity of AtfA to DAG. Meanwhile, the available cellular acyl-CoA and ethanol could also be catalyzed by wax ester synthase activity of AtfA to form FAEE. Therefore, we then assessed the ability of newly discovered WS/DGAT and RoPAP combinations for TAG production. TLC results showed that the combination of tDGAT and RoPAP produced the most abundant TAG, followed by the combinations AtfA and RoPAP, AtfA_co (the codon-optimized AtfA for E. coli) and RoPAP, and Atf8 and RoPAP (Fig. 2b). However, Atf1 or Atf2 combined with RoPAP did not yield detectable TAG, although both Atf1 and Atf2 were responsible for TAG formation in R. opacus PD630, while Atf2 exhibited higher DGAT activity than Atf1 [36, 37]. Unlike AtfA or Sco0958 (WS/DGAT from S. coelicolor), overexpression of a solo tDGAT was enough to accumulate considerable TAGs with a threefold increase in monounsaturated fatty acids in E. coli C41(DE3) [18, 21, 24]. Combined with the data here, tDGAT was highly active in E. coli, and it has the potential for use in the production of functional lipids.
Evaluation of bacterial WS/DGAT and PAP combinations for the production of TAGs in E. coli. AtfA and RoPAP/RjPAP combinations (a) and six different WS/DGATs and RoPAP combinations (b). Cells were cultured in ZYP-5052 auto-induction medium at 37 °C with shaking at 200 rpm for 48 h. Thin-layer chromatography (TLC) was carried out on lipids extracted from 10 mg of dried cells. tDGAT: WS/DGAT from T. curvata; AtfA: WS/DGAT from A. baylyi ADP1; AtfA_co: AtfA codon-optimized for E. coli; Atf8: WS/DGAT from R. jostii RHA1; Atf1/Atf2: WS/DGATs from R. opacus PD630. RoPAP: PAP from R. opacus PD630, RjPAP: PAP from R. jostii RHA1
The protein sequences of all tested WS/DGATs contained the catalytic motif HHxxxDG and two other conserved motifs, as shown in Additional file 1: Figure S1. The predicted three-dimensional structure of tDGAT revealed that it had a longer helical linker between the N- and C-terminal domains. In addition, it has been predicted as the most hydrophobic protein among existing representative WS/DGATs, which led to relatively difficult purification [18].
The results here also indicated that DAG availability was crucial for TAG synthesis and that RoPAP contributed to circumvent this bottleneck. Both RoPAP and RjPAP had six predicted transmembrane helices (http://www.cbs.dtu.dk/services/TMHMM/). Sequence alignment of RoPAP and RjPAP showed two conserved domains PSGH and RxxxxHxxxD, which have also been found in LPPα and LPPβ [21]. In addition, the conserved KxxxxxxRP was present in both RoPAP and RjPAP, but absent in PgpB (Additional file 2: Figure S2A). Expression of a solo RoPAP or RjPAP led to slightly increased TFA content by 14 and 13%, respectively, and the fatty acid profile did not change in either case (Additional file 2: Figure S2B, C).
We also noticed that TAG spots from E. coli cells had much stronger signals when staining with primuline over iodine (Additional file 3: Figure S3). In addition, FFA and DAG were more easily observed by staining with primuline (Additional file 3: Figure S3). Therefore, primuline was used for lipid staining in the following tests.
Acyl-CoA synthetase RofadD1 from R. opaque PD630 enhanced TAG production
To construct an acyl-CoA-dependent TAG pathway in E. coli, it is essential to enhance the acyl-CoA pool in addition to inducing RoPAP for DAG accumulation. Two putative acyl-CoA synthetases were predicted from oleaginous bacterium R. opacus PD630 and were designated as RoFadD1 (EHI43506) and RoFadD2 (AHK27355). Additional expression of RoFadD1 along with tDGAT and RoPAP increased the cellular TAG titer, from 92.5 to 105.9 mg/L (Fig. 3). This suggested that there was a limited cytosolic acyl-CoA pool in engineered E. coli, and thus, activation of FFAs to their corresponding acyl-CoAs by the action of RoFaD1 enhanced TAG synthesis. FFAs were also boosted as the result of RoFadD1 overexpression (Fig. 3). This may be explained by the other important function of acyl-CoA synthetase which is transportation of exogenous fatty acids across the inner membrane [23]. Alternatively, the increased TAGs and FFAs are due to the altered metabolic flux. The sub-location and detailed function of RoFadD1 await further investigations. In contrast, RoFadD2 did not show the ability to increase TAGs when compared with RoFadD1 (Additional file 4: Figure S4).
Coexpression of RoFadD1 and RoTetRs along with tDGAT and RoPAP further increased TAG synthesis. Cells were cultured in ZYP-5052 auto-induction medium at 37 °C with shaking at 200 rpm for 48 h. Thin-layer chromatography (TLC) was carried out on lipids extracted from 5 mg of dried cells. tDGAT, WS/DGAT from T. curvata, RoPAP, PAP from R. opacus PD630. RoFadD1, putative acyl-CoA synthetase from R. opacus PD630. FadR, fatty acid metabolism regulator from E. coli MG1655; RoTetR1/2/3, three putative fatty acid metabolism regulators from R. opacus PD630. All data are the means ± standard deviations from triplicates
Newly discovered fatty acid metabolism regulators from R. opacus PD630 contributed to the increased TAGs
RoTetR1/2/3 from the genome sequence of oleaginous R. opacus PD630 were denoted as three putative fatty acid metabolism regulators, although none of them contributed to the increased TFAs or hydroxyl fatty acids [32]. We tested whether they would enhance TAG synthesis. Results showed that expressing either RoTetR in addition to tDGAT, RoPAP, and RoFadD1, all led to increased TAG accumulation, with values of 149.7 mg/L (RoTetR1), 141.8 mg/L (RoTetR2), and 141.1 mg/L (RoTetR3), respectively (Fig. 3). The universal fatty acid metabolism regulator FadR slightly increased TAGs as well, rising from 105.9 mg/L (without regulator) to 119.5 mg/L (Fig. 3). FFAs were drastically reduced when either RoTetR2 or RoTetR3 was expressed alongside tDGAT, RoPAP and RoFadD1 (Fig. 3), suggesting that most FFAs were converted to their respective acyl-CoAs under the tested conditions. Our previous result indicated that FadR increased the hydroxyl fatty acid production in E. coli, whereas none of the RoTetRs possessed the same ability [32]. Furthermore, only fadR overexpression stimulated the monounsaturated fatty acids C16:1 and C18:1 at the expense of saturated C14:0 in this study, similar to the results reported elsewhere (Additional file 5: Figure S5) [27]. Therefore, we predicted that the regulation mechanism of RoTetRs might be different from that of FadR. Further investigation of the differentially expressed genes regulated by each RoTetR would be helpful for identification of the regulation mechanism.
To alleviate the growth and metabolic burden, four selected genes including tDGAT, RoPAP, RoFadD1, and RoTetR2 were next rearranged into the same pCDFDuet vector to form the plasmid pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2 with the RBS motif upstream of each gene (Additional file 6: Figure S6). By expressing this plasmid in E. coli BL21(DE3) (designated as strain 2119), the TAG titer was further increased to 175.7 mg/L as expected (see results below, Fig. 4). We also observed that lowering the culturing temperature from 37 to 30 °C caused drastically decreased TAGs in strain 2119 (Additional file 7: Figure S7). Because tDGAT was isolated from a thermophilic actinobacterium with an optimal growing temperature of 50 °C [18]. Thus, it was likely that tDGAT exhibited higher activity at 37 °C compared to 30 °C.
Assessment of medium-chain specific TE for diverting MCFA flux into TAG backbones. a MCFAs from the TAG fraction extracted from strain 2119 carrying different TEs. b TAG content and titer of strain 2119 carrying different TEs. c Fatty acid profile of the TAG fraction extracted from engineered strain 2119 carrying different TEs. d Time course of TAG production yielded by strain 2119 carrying pACYCDuet::RcFatB. Thin-layer chromatography (TLC) of lipid extracted from dried cells. e TAG profile and the distribution of C14 in the total TAGs from lipids extracted from strain 2119 and strain 2119 plus pACYCDuet::RcFatB, respectively. Strain 2119: E. coli BL21(DE3) carrying plasmid pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2. 48:1 represents 48 carbons with 1 double bond in three acyl chains. All data are the means ± standard deviations from triplicates
Overexpression of the castor TE gene RcFatB-triggered MCFA flux into TAG backbones
The next step was to selectively release MCFAs from the acyl-ACP pool and incorporate them into TAGs backbone. Six selected MCFA-specific TE genes including CnFatB3, CcFatB1, CpFatB2, ChFatB2, AcTesA’, and RcFatB were first inserted into the low copy number pACYCDuet-1 vector. Each TE was then expressed in strain 2119. Castor RcFatA was used as a control as it exhibited a preference for C16 and C18 instead of MCFAs. TLC combined with GC results showed that the RcFatB combination resulted in the highest MCFA titer (mainly C12:0, C14:0, and C14:1) in the form of TAG, which reached 57.4 mg/L. The increase in MCFA content from 12.7 to 39.0% in the TAGs was mainly at the expense of C16:0 (Fig. 4a, c). On the other hand, additional expression of RcFatB also caused a drop in the total amount of TAGs from 175.7 to 147.4 mg/L (Fig. 4b). Similar to the RcFatB combination, the CpFatB2 combination produced 27.9% MCFAs in the TAG, corresponding to 44.9 mg/L MCFAs in the form of TAG. Again, the total TAGs dropped to 146.2 mg/L in the CpFatB2 combination, similar to that of RcFatB (Fig. 4).
We also observed that the RcFatB combination yielded 140.2 mg/L MCFAs from cell lysate. However, only 41% of the MCFAs were incorporated into TAG backbones, suggesting that a large amount of MCFAs existed in the form of cellular FFA or membrane lipids. Accumulation of high levels of MCFA is known to be toxic to E. coli cells, since MCFAs would be partially incorporated into membrane lipids and would lead to impaired membrane functions [38]. We indeed observed that biomass in the RcFatB or CpFatB2 combinations was decreased with 17.9 and 25.8% reductions observed, respectively. Nevertheless, such a reduction may be an acceptable trade-off, since the value-added TAGs will be produced anyway, and the reductions may be minimized by optimization of nutritional levels. Besides, strategies to prevent the incorporation of MCFA into membrane lipid should be considered in the near future.
RcFatB has been reported as being active on oleoyl-ACP and palmitoyl-ACP in vitro [39]. However, the preference of RcFatB for myristoyl-ACP or lauroyl-ACP substrates was not tested in vitro. In another report, RcFatB was overexpressed in E. coli to produce 2.0 g/L FFAs at 48 h with 40% C14 [40]. The combined expression of RcFatB with other essential type II fatty acid synthesis genes has also been tested in Saccharomyces cerevisiae, leading to the production of 32% C14 fatty acids (20% C14:1 and 12% C14:0) among the TFAs [41]. Combining our results here, we predicted that RcFatB probably would exhibit a myristoyl-ACP preference in vitro, although there was only a trace amount of C14 accumulation in the seed of the RcFatB source plant, castor (Ricinus communis L).
The TAG production from combined expression of RcFatB, tDGAT, RoPAP, RoFadD1, and RoTetR2 during the time course of the assay was then investigated. The TAG content increased to 5.8% (w/w CDW) after culturing for 36 h, while the highest TAG titer achieved was 147.4 mg/L at 48 h (Fig. 4d). The TAGs were accumulated in cells as early as 6 h, although the amount was quite low (Fig. 4d). This was consistent with the previous results, where TAGs were detected after induction of tDGAT expression for only 30 min [18]. Compared to oleaginous bacteria and yeasts, a shorter fermentation time was observed for TAG accumulation in engineered E. coli. This might be beneficial when seeking to produce TAGs in engineered E. coli cells.
Expression of RcFatB also significantly changed the TAG profile (Fig. 4e). With RcFatB, TAG-44:1 and TAG-46:2 accounted for 21 and 19% of the total TAG, respectively, suggesting that 16:1/14:0/14:0 or 16:0/14:1/14:0, and 16:1/16:1/14:0 might be the most abundant TAG classes. This result further confirmed the increased MCFA flux into TAG backbones. In addition, the distribution of MCFAs in TAG species was analyzed. The predominant C14:0 was mainly found in TAG-44:1, representing 32% of the total C14:0 containing TAGs, and indicating that 16:1/14:0/14:0 or 16:0/14:1/14:0 might be the most abundant TAGs for sequestration of C14:0 in the RcFatB combination. In contrast, C14:0 mainly existed in TAG-46:1 without RcFatB expression (Fig. 4f).
We also observed that the MCFAs dropped to control levels (12.5% of the total TAGs) if RcFatB was expressed by the high copy number plasmid pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2/RcFatB, although the total TAG and TFA titers further increased to 234.7 mg/L (7.0% CDW) and 553.2 mg/L (16.5% CDW), respectively (Additional file 6: Figure S6 and Additional file 8: S8). A similar result was observed in the RcFatA combination as well (Additional file 8: Figure S8). The increase in the total TAGs was probably caused by a lower metabolic burden or less MCFA toxicity. However, considering the drastically decreased MCFAs, pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2/RcFatB was not appropriate for MCFA production. Our previous results revealed that expression of CnFatB3, CcFatB1, or CpFatB2 with low copy numbers rather than high copy number plasmids boosted the production of medium-chain hydroxyl fatty acids in E. coli [32]. It is assumed that the binary activities of TE towards both acyl-ACP and acyl-CoA resulted in diverting the acyl pools to FFA. The accumulation of MCFA-enriched TAGs might also depend on the efficiency of the interaction between generated acyl-ACPs and their corresponding applied TEs in engineered E. coli. Therefore, the choice of vector for expressing TEs was also crucial for the production of MCFAs. Further increases in TAG production may be achieved by balancing metabolic fluxes through expression of these genes under optimized promoters and/or RBS.
Collectively, castor RcFatB showed a preference for MCFAs mainly C14 fatty acids in E. coli cells, although C14:0 or C14:1 is not the major fatty acid in castor seed oils [42]. RcFatB may also be considered a good candidate for future production of tailored fatty acids and their derivatives. Furthermore, the results also demonstrated that tDGAT was able to transfer acyl-CoA with tailored fatty acids into TAG backbones, although the efficiency was undetermined.
TAG induction through nutrient-level optimization
We next sought to increase the TAG titer without decreasing the MCFA proportions by optimization of nutrient levels. Auto-induction medium ZYP-5052 was shown to be more suitable for TAG accumulation in E. coli than conventional LB medium with IPTG induction [16]. However, ZYP-5052 was originally designed for high expression of any target protein under the T7 promoter. Thus, it was probably not the best choice for TAG accumulation. Therefore, the ZYP-5052 medium was optimized in this study for improving TAG synthesis (Additional file 10: Table S1). The results here suggested that the ratio of glycerol to N-Z-amine, rather than the absolute concentration of glycerol or N-Z-amine, was crucial towards enhancing both the TAG and MCFA titers (Fig. 5a, b). After optimization, the highest TAG titer was achieved of up to 5.7% (w/w CDW) with 43.8% of MCFAs, corresponding to 399.4 mg/L of total TAGs (Fig. 5d, e). The optimized medium containing 1% N-Z-amine, 5% glycerol, 25 mM (NH4)2SO4, 3 mM MgSO4, 0.5% aspartic acid, and 0.5% serine, and the other ingredients were the same as in ZYP-5052. The TAG titer and content were both elevated as the supplementation with glycerol increased (Fig. 5). Glycerol as a renewable by-product can be channeled into glycolysis to provide a substrate for fatty acids, membrane lipids, and DAG synthesis. More importantly, unlike glucose, the increased glycerol concentration would not prevent induction by lactose in auto-induction media [43]. As to the effect of nitrogen N-Z-amine, despite the fact that increasing nitrogen levels improved the biomass, the TAG content was drastically decreased (Fig. 5a). In addition, neither a change in the NH4+/Mg2+ dosage nor supplementation with aspartic acid and serine caused an obvious TAG change.
TAG induction through nutrient level optimization. Contour profiles of TAG content (a) and TAG titer (b) for strain 2119 harboring pACYCDuet::RcFatB cultured in 16 designed media with different ratios of glycerol and N-Z-amine. Floating cells were observed during the collection of cells (c). Increases in the TAG/MCFA content (d, e) and titer (f) realized from nutrient optimization. The detailed information on designed media is shown in Additional file 10: Table S1. Cells were cultured at 37 °C with shaking at 200 rpm for 48 h. All data are the means ± standard deviations from triplicates
Large amounts of floating cells were also observed during the collection process, especially for cells cultured in the optimized medium. This indicated that these cells had high buoyancy and thus could float in the medium once the culture was settled for 3 h, whereas normal cells settled to the bottom of the tube (Fig. 5c). A similar observation has been described in the oleaginous yeast, Yarrowia lipolytica, which also produces high levels of lipids [44]. The floating characteristics of such engineered cells may facilitate the subsequent purification process.
Small and membrane-bound lipid droplets (LD) were formed in recombinant E. coli
We next investigated whether TAGs rich in MCFAs could be accumulated in the form of LDs. Both N-SIM super-resolution microscopy and TEM analysis clearly showed that small LDs were formed and accumulated in cells after culturing for only 12 h in optimized medium (Fig. 6). After culturing for 24 h or more, large LDs with irregular shapes were observed in cells, suggesting that LDs were fused via an unknown mechanism in late stationary cells (Fig. 6). We also noticed that almost all observed LDs were attached to cell membranes (Fig. 6). Prokaryotic LDs have been considered to be phase-separated organelles in the cytoplasm, and their formation starts with attachment of WS/DGAT to the cytoplasm membrane [45]. Unlike eukaryotic LDs, prokaryotic LDs have been predicted as being surrounded by a monolayer of phospholipids [45, 46]. The observed membrane-bound LDs from engineered E. coli cells were consistent with earlier predictions. In yeast cells, if storage lipids such as TAGs are not generated from inert end product pools, they would be metabolically active [47]. However, this was not the case in the engineered E. coli, as most of the observed LDs were still intact in the lysed cells after culturing for 48 h (Fig. 6b). This characteristic might be beneficial for further purification of prokaryotic LDs.
Observation of LDs in engineered E. coli cells. Cells were observed by transmission electron microscopy (TEM) (a) and N-SIM super-resolution microscope (b). The engineered strain 2119 carrying pACYCDuet::RcFatB cultured in ZYP-5052 auto-induction medium at 37 °C with shaking at 200 rpm for 12, 24, 36 or 48 h. LDs were stained with Nile Red before observing by N-SIM. The scale bar is 1 µm for TEM and 4 µm for N-SIM
Despite the fact that many important proteins contribute to LD formation or stability, for example oleosin, lipid droplet associated protein (LDAP) and caleosin, all of which have been identified and characterized from oil plants, few functional proteins have been revealed from oleaginous bacteria [48]. However, one such representative protein is TadA from R. opacus PD630, and mutation of TadA has led to 30–40% less TAGs and smaller LDs when compared to the parental strain [49]. However, we did not find any homologues of TadA in E. coli and overexpression of TadA dramatically decreased the TAG titer in the engineered TAG-producing strain 2119 (Additional file 9: Figure S9). Furthermore, the Ro02104 protein isolated from R. jostii RHA1 also plays a role in LD dynamics [50]. Recently, an LD-associated protein MLDS from R. jostii RHA1 was demonstrated to mediate binding of LDs to genomic DNA and thus enhance the cell survival under stress [51]. Whether the LDs generated by engineered E. coli could exert similar roles is unknown. Nevertheless, the formation of LDs did not cause obvious cell lysis or impair cell growth in our study. It is presumed that identification of the mechanism involved in prokaryotic LD formation and characterization of LD-associated proteins may facilitate the stability of LDs or further enhance the TAG titer in engineered strains. Collectively, TAGs with tailored fatty acids can be stored in the LDs of engineered E. coli. Such prokaryotic LDs may be exploited for various biotechnology applications due to their simple yet robust structures, as well as their amenability to extraction.
Conclusions
In conclusion, we reconstructed an acyl-CoA-dependent pathway in E. coli by introducing four heterologous genes including tDGAT, RoPAP, RoFadD1, and RoTetR2 to produce high levels of TAG. Co-expression of castor RcFatB with the above genes created MCFA substrates available for TAG assembly. The titers of TAGs rich in MCFAs reached 399.4 mg/L, attaining their highest levels under shake flask conditions. In addition, membrane-bound LDs were formed in engineered E. coli. This illustrated that E. coli as a prokaryotic cell factory has the potential to produce value-added TAG.
Methods
Chemicals, enzymes, and kits
Chemicals including primuline, hexane, acetone, acetic acid, and diethyl ester were purchased from Sigma-Aldrich (St. Louis, USA) or Sinopharm (Shanghai, China). Methanol, chloroform, Phusion polymerase, restriction enzymes, T4 DNA ligase, DNase I, and RevertAid First Strand cDNA Synthesis Kit were purchased from Thermo Fisher Scientific (Beverly, USA). Plasmid Miniprep Kit, Total DNA Extraction Kit, PCR Purification Kit, and Gel Extraction Kit were purchased from TianGen (Beijing, China). RNA Protect Reagent and RNeasy Mini Kit were purchased from Qiagen (Hilden, Germany). Power SYBR® Green PCR Master Mix was from Applied Biosystems (Massachusetts, USA). FFA, DAG, TAG, or FAEE standards were purchased from Nu-Chek (Elysian, USA) or Larodan (Stockholm, Sweden).
Strains, media, and growth conditions
Escherichia coli DH5α was used for plasmid construction and propagation. E. coli BL21(DE3) was used for gene expressing and TAG production. Cells were cultivated in Luria–Bertani medium (LB, 1% (w/v) tryptone, 0.5% (w/v) yeast extract, and 1% (w/v) NaCl) at 37 °C with shaking at 200 rpm for cell inoculation or plasmid propagation. Appropriate antibiotics were added at the following concentrations: kanamycin 50 mg/L, ampicillin 100 mg/L, spectinomycin 50 mg/L, or chloramphenicol 34 mg/L, respectively. Auto-induction medium ZYP-5052 with appropriate antibiotics was mainly used for TAG production [43]. For time-course experiment, cells were collected after culturing in ZYP-5052 at 37 °C with shaking at 200 rpm for 6, 12, 24, 36, and 48 h, respectively.
For nutrient optimization, 16 designed media were tested for TAG production. The detailed information of the designed media is listed in Additional file 10: Table S1. Cells were cultured at 37 °C with shaking at 200 rpm for 48 h.
Plasmid construction and gene expression
All constructed strains and plasmids used in this study are summarized in Additional file 11: Table S2. Oligonucleotides used as PCR primers are listed in Additional file 13: Table S3. PAP encoding genes RoPAP (WP_005246202) from R. opacus PD630 and RjPAP (WP_011593404) from R. Jostii RHA1 were chemically synthesized and inserted into pCDFDuet-1, respectively. Then, each gene was subcloned into the downstream of atfA (AAO17391, from A. baylyi ADP1) flanked by a Ribosome-Binding Site (RBS) sequence (AAGGAG). Other WS/DGAT encoding genes include atf1 (EHI42943) and atf2 (EHI41112) from Rhodococcus opacus PD630, atf8 (ACY38595) from R. jostii RHA1, tDGAT (WP_012854133) from Thermomonospora curvata DSM 43183, and atfA_co (atfA was codon optimized for E. coli expression). Each above WS/DGAT encoding gene was chemically synthesized and subcloned into upstream of RoPAP flanked by a RBS sequence, located in the EcoRI/NotI sites of pCDFDuet-1. Acyl-CoA synthetase genes RoFadD1 (EHI43506) or RoFadD2 (AHK27355), from R. opacus PD630, were then inserted into the NdeI/XhoI located at the second multiple cloning sites (MCS2) of pCDFDuet-1. Fatty acid metabolism regulators fadR from E. coli, or RoTetR1, RoTetR2, and RoTetR3 from R. opaque PD630 were subcloned into pET28a in our previous work [32]. For assembling multiple genes into one vector, tDGAT and RoPAP, flanked by RBS were introduced into EcoRI/NotI sites of pCDFDuet-1, while RoFadD1 and RoTetR2 flanked by RBS were inserted into NdeI/XhoI sites, generating the plasmid pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2. The detailed information on constructed plasmids is shown in Additional file 6: Figure S6.
TE genes RcFatA (NP_001310680) and RcFatB (NP_001310677) from Ricinus communis, ChFatB2 (AAC49269) from C. hookeriana, AcTesA’ from A. baylyi were all codon-optimized, chemically synthesized, and inserted into pACYCDuet-1, respectively. Three other plant TE genes, CnFatB3, CcFatB1, and CpFatB2, previously constructed in pACYCDuet-1 vector [32], were also used in this study. The gene RcFatA or RcFatB with RBS at 5′-end was also inserted into pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2 via seamless assembly technology to generate pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2/RcFatA or pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2/RcFatB, respectively (Additional file 6: Figure S6).
Lipid analysis
Escherichia coli cells were cultured each in 50 mL medium, and the mass of the freeze-dried cells was measured. Total lipids were isolated from two and half or five milligram of the freeze-dried E. coli cells according to a previous method [16]. The extracted lipids were separated by thin-layer chromatography (TLC) using a solvent system (hexane:diethyl ester:acetic acid = 70:30:1), and visualized by spraying with 0.01% primuline dissolved in 80% acetone. The bands corresponding to FFA and TAG were recovered from TLC plates and methylated as previously described [1]. The resulting fatty acid methyl esters (FAMEs) were then analyzed using a 7890A gas chromatography (GC, Agilent technologies, USA) equipped with a flame ionization detector (FID) and an HP-FFAP column (30 m × 250 µm i.d. × 0.25 µm thickness). For quantification, either 10 µg of C15:0-TAG and 5 µg of C15:0-FFA (for low levels of TAG or FFA) or 20 µg of C15:0-TAG and 10 µg of C15:0-FFA were added into samples as internal standards before lipid extraction, respectively. Lipids from 2 mg dried cells were directly methylated to FAMEs and analyzed by GC as described above, to quantify cellular TFA.
Shotgun lipidomic analysis of TAG profile was done by liquid chromatography–mass spectrometry (LC–MS) [52]. Briefly, a hybrid triple quadrupole/linear IT mass spectrometer, API 4000 Q-Trap (AB SCIEX, Foster City, CA, USA), operating under an Analyst software system was utilized here. The heater temperature along the ion transfer capillary was maintained at 300 °C. The shelter gas (nitrogen) pressure was 344.75 kPa. The diluted lipid (0.01–0.06 mg/mL) was directly infused into the ESI source at a flow rate of 0.02 mL/min. TAG profile was analyzed with tools at the website: http://www.lipidmaps.org.
Microscopy analysis
For transmission electron microscopy (TEM) imaging, cells were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer pH7.4 under vacuum for 24 h. For super-resolution microscopy observation, aliquots (200 µL) of cell cultures grown for 12, 24, 36, or 48 h were harvested, washed and dissolved in phosphate-buffered saline (PBS), and then stained with 2 µL of Nile Red (1 mg/mL in acetone stock) for 30 min in the dark. Stained cells were washed three times with PBS and then suspended in 100 µL PBS. Samples were observed immediately using a Structured Illumination Microscope (N-SIM, Nikon, Japan) equipped with an EMCCD camera (iXon DU-897, Andor, Northern Ireland) and a 100 × 1.49 NA TIRF objective (CFI Apo TIRF, Nikon, Japan). The fluorescence was excited with a 561 nm laser and detected by a 570–640 nm bandpass emission filter. Images were captured in multiple phases and angles of the illumination pattern and then directly reconstructed with the Nikon NIS-Elements Ar software. The final pixel size of SIM image was 30 nm.
Abbreviations
- MCFA:
-
medium-chain fatty acids
- CDW:
-
cell dry weight
- DAG:
-
diacylglycerol
- WS/DGAT:
-
wax ester synthase/acyl-Coenzyme A:diacylglycerol acyltransferase
- FAME:
-
fatty acid methyl ester
- FFA:
-
free fatty acid
- TFA:
-
total fatty acid
- SCO:
-
single-cell oil
- TAG:
-
triacylglycerol
- acyl-ACP:
-
acyl-acyl carrier protein
- TE:
-
acyl-ACP thioesterase
- TLC:
-
thin-layer chromatography
- FAEE:
-
fatty acid ethyl ester
- LD:
-
lipid droplet
References
Vanhercke T, Divi UK, El Tahchy A, Liu Q, Mitchell M, Taylor MC, Eastmond PJ, Bryant F, Mechanicos A, Blundell C, et al. Step changes in leaf oil accumulation via iterative metabolic engineering. Metab Eng. 2017;39:237–46.
Zargar A, Bailey CB, Haushalter RW, Eiben CB, Katz L, Keasling JD. Leveraging microbial biosynthetic pathways for the generation of ‘drop-in’ biofuels. Curr Opin Biotechnol. 2017;45:156–63.
Kallio P, Pasztor A, Akhtar MK, Jones PR. Renewable jet fuel. Curr Opin Biotechnol. 2014;26:50–5.
Gutierrez-Garcia AG, Contreras CM, Diaz-Marte C. Myristic acid in amniotic fluid produces appetitive responses in human newborns. Early Hum Dev. 2017;115:32–7.
Contreras CM, Rodriguez-Landa JF, Garcia-Rios RI, Cueto-Escobedo J, Guillen-Ruiz G, Bernal-Morales B. Myristic acid produces anxiolytic-like effects in Wistar rats in the elevated plus maze. Biomed Res Int. 2014;2014:492141.
Jing F, Cantu DC, Tvaruzkova J, Chipman JP, Nikolau BJ, Yandeau-Nelson MD, Reilly PJ. Phylogenetic and experimental characterization of an acyl-ACP thioesterase family reveals significant diversity in enzymatic specificity and activity. BMC Biochem. 2011;12:44.
Iskandarov U, Silva JE, Kim HJ, Andersson M, Cahoon RE, Mockaitis K, Cahoon EB. A specialized diacylglycerol acyltransferase contributes to the extreme medium-chain fatty acid content of Cuphea seed oil. Plant Physiol. 2017;174:97–109.
Reynolds KB, Taylor MC, Cullerne DP, Blanchard CL, Wood CC, Singh SP, Petrie JR. A reconfigured Kennedy pathway which promotes efficient accumulation of medium-chain fatty acids in leaf oils. Plant Biotechnol J. 2017;15:1397–408.
Xu P, Qiao K, Ahn WS, Stephanopoulos G. Engineering Yarrowia lipolytica as a platform for synthesis of drop-in transportation fuels and oleochemicals. Proc Natl Acad Sci USA. 2016;113:10848–53.
Wu J, Zhang X, Xia X, Dong M. A systematic optimization of medium chain fatty acid biosynthesis via the reverse beta-oxidation cycle in Escherichia coli. Metab Eng. 2017;41:115–24.
Wang C, Pfleger BF, Kim SW. Reassessing Escherichia coli as a cell factory for biofuel production. Curr Opin Biotechnol. 2017;45:92–103.
Parsons JB, Rock CO. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res. 2013;52:249–76.
Rottig A, Steinbuchel A. Acyltransferases in bacteria. Microbiol Mol Biol Rev. 2013;77:277–321.
Theodoulou FL, Eastmond PJ. Seed storage oil catabolism: a story of give and take. Curr Opin Plant Biol. 2012;15:322–8.
Rottig A, Strittmatter CS, Schauer J, Hiessl S, Poehlein A, Daniel R, Steinbuchel A. Role of wax ester synthase/acyl Coenzyme A: diacylglycerol acyltransferase in oleaginous Streptomyces sp. strain G25. Appl Environ Microbiol. 2016;82:5969–81.
Rottig A, Zurek PJ, Steinbuchel A. Assessment of bacterial acyltransferases for an efficient lipid production in metabolically engineered strains of E. coli. Metab Eng. 2015;32:195–206.
Amara S, Seghezzi N, Otani H, Diaz-Salazar C, Liu J, Eltis LD. Characterization of key triacylglycerol biosynthesis processes in Rhodococci. Sci Rep. 2016;6:24985.
Lazaro B, Villa JA, Santin O, Cabezas M, Milagre CDF, de la Cruz F, Moncalian G. Heterologous expression of a thermophilic diacylglycerol acyltransferase triggers triglyceride accumulation in Escherichia coli. PLoS ONE. 2017;12:e0176520.
Jin HH, Jiang JG. Phosphatidic acid phosphatase and diacylglycerol acyltransferase: potential targets for metabolic engineering of microorganism oil. J Agric Food Chem. 2015;63:3067–77.
Hernandez MA, Comba S, Arabolaza A, Gramajo H, Alvarez HM. Overexpression of a phosphatidic acid phosphatase type 2 leads to an increase in triacylglycerol production in oleaginous Rhodococcus strains. Appl Microbiol Biotechnol. 2015;99:2191–207.
Comba S, Menendez-Bravo S, Arabolaza A, Gramajo H. Identification and physiological characterization of phosphatidic acid phosphatase enzymes involved in triacylglycerol biosynthesis in Streptomyces coelicolor. Microb Cell Factories. 2013;12:9.
Touze T, Blanot D, Mengin-Lecreulx D. Substrate specificity and membrane topology of Escherichia coli PgpB, an undecaprenyl pyrophosphate phosphatase. J Biol Chem. 2008;283:16573–83.
Fujita Y, Matsuoka H, Hirooka K. Regulation of fatty acid metabolism in bacteria. Mol Microbiol. 2007;66:829–39.
Janssen HJ, Steinbuchel A. Production of triacylglycerols in Escherichia coli by deletion of the diacylglycerol kinase gene and heterologous overexpression of atfA from Acinetobacter baylyi ADP1. Appl Microbiol Biotechnol. 2014;98:1913–24.
Guo X, Jiang M, Wan X, Hu C, Gong Y. Identification and biochemical characterization of five long-chain acyl-coenzyme A synthetases from the diatom Phaeodactylum tricornutum. Plant Physiol Biochem. 2014;74:33–41.
Jessen D, Roth C, Wiermer M, Fulda M. Two activities of long-chain acyl-coenzyme A synthetase are involved in lipid trafficking between the endoplasmic reticulum and the plastid in Arabidopsis. Plant Physiol. 2015;167:351–66.
Zhang F, Ouellet M, Batth TS, Adams PD, Petzold CJ, Mukhopadhyay A, Keasling JD. Enhancing fatty acid production by the expression of the regulatory transcription factor FadR. Metab Eng. 2012;14:653–60.
Comba S, Sabatini M, Menendez-Bravo S, Arabolaza A, Gramajo H. Engineering a Streptomyces coelicolor biosynthesis pathway into Escherichia coli for high yield triglyceride production. Biotechnol Biofuels. 2014;7:172.
My L, Ghandour Achkar N, Viala JP, Bouveret E. Reassessment of the genetic regulation of fatty acid synthesis in Escherichia coli: global Positive control by the dual functional regulator FadR. J Bacteriol. 2015;197:1862–72.
Lennen RM, Pfleger BF. Engineering Escherichia coli to synthesize free fatty acids. Trends Biotechnol. 2012;30:659–67.
Li M, Zhang X, Agrawal A, San KY. Effect of acetate formation pathway and long chain fatty acid CoA-ligase on the free fatty acid production in E. coli expressing acy-ACP thioesterase from Ricinus communis. Metab Eng. 2012;14:380–7.
Xiao K, Yue XH, Chen WC, Zhou XR, Wang L, Xu L, Huang FH, Wan X. Metabolic engineering for enhanced medium chain omega hydroxy fatty acid production in Escherichia coli. Front Microbiol. 2018;9:139.
Dehesh K, Jones A, Knutzon DS, Voelker TA. Production of high levels of 8:0 and 10:0 fatty acids in transgenic canola by overexpression of ChFatB2, a thioesterase cDNA from Cuphea hookeriana. Plant J. 1996;9:167–72.
Kim HJ, Silva JE, Vu HS, Mockaitis K, Nam JW, Cahoon EB. Toward production of jet fuel functionality in oilseeds: identification of FatB acyl-acyl carrier protein thioesterases and evaluation of combinatorial expression strategies in Camelina seeds. J Exp Bot. 2015;66:4251–65.
Ukey R, Holmes WE, Bajpai R, Chistoserdov AY. Evaluation of thioesterases from Acinetobacter baylyi for production of free fatty acids. Can J Microbiol. 2017;63:321–9.
Hernandez MA, Arabolaza A, Rodriguez E, Gramajo H, Alvarez HM. The atf2 gene is involved in triacylglycerol biosynthesis and accumulation in the oleaginous Rhodococcus opacus PD630. Appl Microbiol Biotechnol. 2013;97:2119–30.
Alvarez AF, Alvarez HM, Kalscheuer R, Waltermann M, Steinbuchel A. Cloning and characterization of a gene involved in triacylglycerol biosynthesis and identification of additional homologous genes in the oleaginous bacterium Rhodococcus opacus PD630. Microbiology. 2008;154:2327–35.
Sherkhanov S, Korman TP, Bowie JU. Improving the tolerance of Escherichia coli to medium-chain fatty acid production. Metab Eng. 2014;25:1–7.
Sánchez-García A, Moreno-Pérez AJ, Muro-Pastor AM, Salas JJ, Garcés R, Martínez-Force E. Acyl-ACP thioesterases from castor (Ricinus communis L.): an enzymatic system appropriate for high rates of oil synthesis and accumulation. Phytochemistry. 2010;71:860–9.
Zhang X, Li M, Agrawal A, San KY. Efficient free fatty acid production in Escherichia coli using plant acyl-ACP thioesterases. Metab Eng. 2011;13:713–22.
Fernandez-Moya R, Leber C, Cardenas J, Da Silva NA. Functional replacement of the Saccharomyces cerevisiae fatty acid synthase with a bacterial type II system allows flexible product profiles. Biotechnol Bioeng. 2015;112:2618–23.
Chan AP, Crabtree J, Zhao Q, Lorenzi H, Orvis J, Puiu D, Melake-Berhan A, Jones KM, Redman J, Chen G, et al. Draft genome sequence of the oilseed species Ricinus communis. Nat Biotechnol. 2010;28:951–6.
Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–34.
Liu L, Pan A, Spofford C, Zhou N, Alper HS. An evolutionary metabolic engineering approach for enhancing lipogenesis in Yarrowia lipolytica. Metab Eng. 2015;29:36–45.
Waltermann M, Hinz A, Robenek H, Troyer D, Reichelt R, Malkus U, Galla HJ, Kalscheuer R, Stoveken T, von Landenberg P, Steinbuchel A. Mechanism of lipid-body formation in prokaryotes: how bacteria fatten up. Mol Microbiol. 2005;55:750–63.
Chapman KD, Dyer JM, Mullen RT. Biogenesis and functions of lipid droplets in plants: thematic review series: lipid droplet synthesis and metabolism: from yeast to man. J Lipid Res. 2012;53:215–26.
Rajakumari S, Grillitsch K, Daum G. Synthesis and turnover of non-polar lipids in yeast. Prog Lipid Res. 2008;47:157–71.
Gidda SK, Park S, Pyc M. Lipid droplet-associated proteins (LDAPs) are required for the dynamic regulation of neutral lipid compartmentation in plant cells. Plant Physiol. 2016;170:2052–71.
MacEachran DP, Prophete ME, Sinskey AJ. The Rhodococcus opacus PD630 heparin-binding hemagglutinin homolog TadA mediates lipid body formation. Appl Environ Microbiol. 2010;76:7217–25.
Ding Y, Yang L, Zhang S, Wang Y, Du Y, Pu J, Peng G, Chen Y, Zhang H, Yu J, et al. Identification of the major functional proteins of prokaryotic lipid droplets. J Lipid Res. 2012;53:399–411.
Zhang C, Yang L, Ding Y, Wang Y, Lan L, Ma Q, Chi X, Wei P, Zhao Y, Steinbuchel A, et al. Bacterial lipid droplets bind to DNA via an intermediary protein that enhances survival under stress. Nat Commun. 2017;8:15979.
Gross RW, Han X. Shotgun lipidomics of neutral lipids as an enabling technology for elucidation of lipid-related diseases. Am J Physiol Endocrinol Metab. 2009;297:E297–303.
Authors’ contributions
LW, XZ, SS, FH, and XW conceived this project. XL, LW, and ZH carried out experimental work. LW, WC, and XW analyzed and interpreted the data. XZ, SS, and XW wrote the paper. All authors assisted in the process. All authors read and approved the final manuscript.
Acknowledgements
We are grateful to Ling Luo for providing help with electronic artwork.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in this article and its additional files.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was supported by Chinese Academy of Agricultural Sciences (Grant Number CAAS-ASTIP-2013-OCRI) and Ministry of Science and Technology of the People’s Republic of China (Grant Number 2016YFD0501209).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Additional files
Additional file 1: Figure S1.
Sequence alignment of five bacterial WS/DGATs. tDGAT: WS/DGAT from T. curvata; AtfA: WS/DGAT from A. baylyi ADP1; Atf8: WS/DGAT from R. jostii RHA1; Atf1/Atf2: WS/DGATs from R. opacus PD630. The predicted catalytic motif “HHxxxDG”, motif I “PLW” and motif II “ND” were indicated by arrows.
Additional file 2: Figure S2.
Comparison of three bacterial phosphatidic acid phosphatases (PAPs). A, sequence alignment of bacterial PAPs. B, TFAs produced by the engineered cells harboring empty vector pCDFDuet-1, RjPAP or RoPAP, respectively. C, fatty acid profile of the cellular fatty acids from the engineered E. coli. PgpB, PAP from E. coli MG1655; RoPAP, PAP from R. opacus PD630; RjPAP, PAP from R. jostii RHA1. All data are the means ± standard deviations from triplicates.
Additional file 3: Figure S3.
Comparison of primuline (A) and iodine (B) staining of neutral lipids extracted from the engineered E. coli cells.
Additional file 4: Figure S4.
A putative acyl-CoA synthetase RoFadD2 from R. opacus PD630 did not increase TAG. Cells were cultured in ZYP-5052 auto-induction medium at 37 °C with shaking at 200 rpm for 48 hr. Lipids extracted from 2.5 mg dried cells were loaded on TLC plate.
Additional file 5: Figure S5.
Fatty acid profile of extracted TAGs from engineered E. coli BL21(DE3) strains harboring different gene combinations. tDGAT: WS/DGAT from T. curvata; RoPAP: PAP from R. opacus PD630; RoFadD1: putative acyl-CoA synthetase from R. opacus PD630; RoTetR1/2/3: three putative fatty acid metabolism regulators from R. opacus PD630; FadR: fatty acid metabolism regulator from E. coli MG1655. All data are the means ± standard deviations from triplicates.
Additional file 6: Figure S6.
The detailed information on representative constructed plasmids.
Additional file 7: Figure S7.
Effect of the incubation temperature on TAG synthesis. Strain 2119 was cultured in ZYP-5052 auto-induction mediumat 30 or 37 °C with shaking at 200 rpm for 48 hr. Strain 2119, E.coli BL21(DE3) harboring pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2.
Additional file 8: Figure S8.
One-vector or two-vector system used for the expression of castor acyl-ACP thioesterase genes RcFatA and RcFatB. The total TAG content and titer (A), or the TFA content and titer (B) from four engineered E.coli strains. 2119+pACY-RcFatA, strain 2119 harboring pACYCDuet::RcFatA; 2119+pACY-RcFatB, strain 2119 harboring pACYCDuet::RcFatB. 2119-RcFatA, E. coli BL21(DE3) harboring pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2/RcFatA. 2119-RcFatB, E.coli BL21(DE3) harboring pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2/RcFatB. Strain 2119, E.coli BL21(DE3) harboring pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2. All data are the means ± standard deviations from triplicates.
Additional file 9: Figure S9.
Overexpression of an LD-associated protein-encoding gene TadA decreased the TAG titer in strain 2119. Cells were cultured in ZYP-5052 auto-induction medium at 37°C with shaking at 200 rpm for 48 hr. Lipids extracted from 5 mg dried cells were loaded on TLC plate. Strain 2119, E.coli BL21(DE3) harboring pCDFDuet::tDGAT/RoPAP::RoFadD1/RoTetR2.
Additional file 10: Table S1.
16 media were designed based on the ZYP-5052 auto-induction medium for improving the accumulation of TAGs rich in MCFAs.
Additional file 11: Table S2.
Strains and plasmids used in this study.
Additional file 12: Table S3.
Primers used in this study.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Xu, L., Wang, L., Zhou, XR. et al. Stepwise metabolic engineering of Escherichia coli to produce triacylglycerol rich in medium-chain fatty acids. Biotechnol Biofuels 11, 177 (2018). https://doi.org/10.1186/s13068-018-1177-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13068-018-1177-x