Abstract
Background
Hormones impact breast tissue proliferation. Studies investigating the associations of circulating hormone levels with mammographic breast density have reported conflicting results. Due to the limited number of studies, we investigated the associations of hormone gene expression as well as their downstream mediators within the plasma with mammographic breast density in postmenopausal women.
Methods
We recruited postmenopausal women at their annual screening mammogram at Washington University School of Medicine, St. Louis. We used the NanoString nCounter platform to quantify gene expression of hormones (prolactin, progesterone receptor (PGR), estrogen receptor 1 (ESR1), signal transducer and activator of transcription (STAT1 and STAT5), and receptor activator of nuclear factor-kB (RANK) pathway markers (RANK, RANKL, osteoprotegerin, TNFRSF18, and TNFRSF13B) in plasma. We used Volpara to measure volumetric percent density, dense volume, and non-dense volume. Linear regression models, adjusted for confounders, were used to evaluate associations between gene expression (linear fold change) and mammographic breast density.
Results
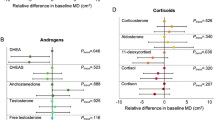
One unit increase in ESR1, RANK, and TNFRSF18 gene expression was associated with 8% (95% CI 0–15%, p value = 0.05), 10% (95% CI 0–20%, p value = 0.04) and % (95% CI 0–9%, p value = 0.04) higher volumetric percent density, respectively. There were no associations between gene expression of other markers and volumetric percent density. One unit increase in osteoprotegerin and PGR gene expression was associated with 12% (95% CI 4–19%, p value = 0.003) and 7% (95% CI 0–13%, p value = 0.04) lower non-dense volume, respectively.
Conclusion
These findings provide new insight on the associations of plasma hormonal and RANK pathway gene expression with mammographic breast density in postmenopausal women and require confirmation in other studies.
Similar content being viewed by others
Background
Mammographic breast density (MBD), a strong risk factor for breast cancer, reflects the amount of epithelial and stromal tissues relative to adipose tissue in the breast [1]. Fat appears darker than epithelium and stroma on a mammogram. Women with greater than 75% density on a mammogram have a 4–6 times greater risk of developing breast cancer compared to women with less than 5% density [2]. The Interventional Breast Cancer Intervention Study demonstrated that a pharmacologically induced 10% decrease in MBD over time is clinically meaningful in the context of breast cancer risk reduction [3].
MBD declines post menopause as endogenous hormone levels decline, indicating an association with hormones and age [4]. MBD also increases with menopausal hormone therapy use [5,6,7,8], and stopping hormone therapy conversely reverts MBD to prior levels [9]. Nevertheless, studies investigating the associations of circulating hormone levels with MBD report conflicting results [10,11,12,13,14,15,16,17]. Few studies have addressed the genomic signatures of MBD or investigated how hormone gene expression (e.g., progesterone and prolactin) in tissue or blood may be associated with MBD [18,19,20]. Gene expression may capture transcriptional changes associated with MBD and could help identify biomarkers of MBD. Breast tissue estrogen receptor 1 (ESR1) gene expression was shown to be negatively associated with percent density in postmenopausal women [21].
We have reported that the receptor activator of nuclear factor kappa-Β ligand (RANKL) gene expression is positively associated with MBD in premenopausal women [22], but there are no data on the associations of plasma RANKL and other tumor necrosis factor receptor superfamily members (e.g., TNFRSF18 and TNFRSF13B) gene expression with MBD in postmenopausal women. Preclinical studies have shown that RANK/RANKL signaling is the major mediator of progesterone-induced mammary epithelial proliferation and expansion of mammary stem cells [23]. Progesterone and prolactin also upregulate RANKL expression [24, 25] and interact with the RANK/RANKL pathway through signal transducer and activator of transcription (STAT) signaling [26, 27]. Despite the extensive crosstalk between the hormones and the RANK pathway identified in preclinical studies, their correlations have not yet been evaluated in population-based studies.
Our objectives in this study are twofold: investigate for the first time the (1) associations of plasma hormone and RANK pathway gene expression with volumetric measures of MBD in postmenopausal women; (2) correlations between hormone and RANK pathway gene expression. Study findings should provide new insight into gene expression profiles that may influence MBD in postmenopausal women.
Methods
Study population
We recruited 400 postmenopausal women during annual routine screening mammography at the Joanne Knight Breast Health Center (BHC) at the Siteman Cancer Center at Washington University School of Medicine, St. Louis, MO between October 2017 and September 2018. Complete data on gene expression and MBD were available and analyzed for 368 women.
Women were eligible to participate in the study if they were: (1) aged 50–64 years; (2) postmenopausal; (3) able to comply with all required study procedures and schedule, including the provision of blood samples at the time of enrollment. Exclusion criteria included: (1) cancer history; (2) history of breast augmentation, reduction, or implants; (3) history of denosumab (a monoclonal antibody that binds RANKL) use in the previous 6 months; (4) history of selective estrogen receptor modulators use in the previous 6 months. We used a modification of the National Comprehensive Cancer Network definition [28], which does not require the measurement of plasma hormone levels to define postmenopausal status. A woman was considered postmenopausal if she had a prior bilateral oophorectomy, was age 60 or older, or if under age 60, had been amenorrheic for at least 12 months.
On the day of the screening mammogram, study participants completed a blood draw and responded to a questionnaire on breast cancer risk factors. Blood samples were processed and stored at − 80 °C within 60 min of collection. A study coordinator measured study participants’ heights using a stadiometer and weights using the OMRON Full Body Sensor Body Composition Monitor and Scale model HBF-514FC. Body mass index (BMI) was derived by dividing current weight (kg) by height (m) squared (kg/m2). Approval for the study was granted by the Institutional Review Board at Washington University School of Medicine, St. Louis, MO. All participants provided written informed consent to participate in the study.
Mammographic breast density assessment
We used Volpara [version 1.5, (Matakina Technology Limited, Wellington, New Zealand)] to obtain automated, objective MBD measurements. Volpara density measurements are highly reproducible [29,30,31]. Volpara uses a relative physics approach and a computerized algorithm that compares. X-ray attenuation at each pixel to a reference pixel within the breast that is assumed to comprise all adipose/non-dense tissue. Using known X-ray attenuation coefficients for fibroglandular/dense and non-dense tissue, Volpara can then estimate the relative thickness of dense and non-dense tissue at each pixel in the image. As the pixel dimensions are known, these thickness estimates can then be converted to volumes and summed across the breast to determine the absolute volumes of dense volume (DV, cm3), and non-dense volume (NDV, cm3) in cubic centimeters. i.e., tissue volume at each pixel = tissue thickness × pixel width × pixel length. The total breast volume is determined using Volpara's proprietary segmentation of the breast and model of the breast edge under compression, and the reported compressed breast thickness. The volumetric breast density (%), can then be determined by taking the ratio of the absolute dense volume to the total breast volume, expressed as a percentage. i.e., Volumetric breast density (VPD, %), % = (volume fibroglandular tissue/volume breast) × 100. In comparison with Breast Imaging Reporting and Data System (BI-RADS) fifth edition, Volpara VPD ranges from 0.5 to 34.5%, which translate to: < 3.5% (a, almost entirely fatty breasts); ≥ 3.5–< 7.5% (b, scattered areas of fibroglandular density); ≥ 7.5–< 15.5% (c, heterogeneously dense breasts); ≥ 15.5 (d, extremely dense breasts).
Plasma gene expression
We performed RNA profiling to quantify gene expression in the plasma, not in the breast tissue to gain further knowledge into how these biomarkers are associated with MBD outside that gleamed from circulating protein levels alone. While mRNA expression and protein levels are correlated across cell lines [32, 33], many factors, including post-translational stability influence circulating protein levels, hence, the correlations of mRNA and their circulating protein levels may be weak [34].
We designed a custom NanoString nCounter codeset for quantitative RNA profiling of the following genes: (1) hormones: prolactin (PRL), progesterone receptor (PGR), estrogen receptor 1 (ESR1), signal transducer and activator of transcription 1 (STAT1), and signal transducer and activator of transcription 5 (STAT5); (2) RANK pathway: RANK, RANKL, tumor necrosis factor receptor superfamily member 13B (TNFRSF13B), tumor necrosis factor receptor superfamily member 18 (TNFRSF18) and osteoprotegerin (OPG). We selected these genes based on data from preclinical studies suggesting crosstalk between the RANK pathway markers (RANK, RANKL, OPG) [35, 36] and specific hormone signaling (PRL, ESR1, PGR, STAT1, and STAT5) [27, 37,38,39]. TNFRSF13 and TNFRSF18 are also RANK pathway markers that could have biological relevance, but there is limited or no data on their associations with MBD. Thus, we designed targets for those genes as well with NanoString.
RNA profiling for gene expression was performed at the McDonnell Genome Institute, Washington University School of Medicine in St. Louis. Gene expression levels were measured in plasma RNA isolated, using the NanoString “nCounter XT Codeset Gene Expression Assays” protocol (NanoString Technologies, Seattle, WA, USA). Quality control was performed as recommended by the manufacturer. This NanoString protocol has been validated extensively in tissue [40,41,42,43] and blood [44, 45].
Plasma samples were processed according to the manufacturer's recommendations. Following hybridization, samples were processed on the NanoString Prep Station where they were purified and immobilized on a sample cartridge for data collection. The output for each sample was imported into nSolver Analysis Software for Quality Control and analysis. Binding densities ranged from 0.09 to 0.34. Digital transcript counts from the NanoString nCounter assay were normalized using the housekeeping genes following the manufacturer’s guidelines.
Statistical analyses
Statistical analyses were performed with the NanoString nSolver Analysis System 4.0 (NanoString Technologies) using the Advanced Analysis package 2.0 and its custom analysis pipeline. VPD, DV, and NDV were all log-transformed to ensure the normality of the residuals. All analyses were performed on curated log2 transformed normalized counts. We evaluated correlations between the genes as well as between the genes and age and BMI using Pearson correlation (r). In addition, we evaluated correlations of the genes adjusted for age and BMI. We also performed correlation analysis in a subset of our study participants (82 women with dense breasts) who had both circulating RANK, RANKL, and OPG and mRNA gene expression data. Genes were tested for differential expression to MBD and adjusted for the following confounding variables: race (Non-Hispanic white/African-American/Others), current age (continuous, years), BMI (continuous, kg/m2), age at menarche (continuous), menopausal hormone therapy use (ever/never), parity, and age at first birth (continuous). For each gene expression, a single linear regression was fit using all selected variables to predict expression. The fold change is then estimated using a simplified negative binomial model, presented here as ‘linear fold change.’ The 95% confidence interval for the linear fold change is also presented, along with a p value. A value of p < 0.05 was considered statistically significant. These linear fold changes are herein discussed in terms of percentage increases or decreases such that a linear fold change of 1.04 corresponds to a 4% increase in MBD, and a 0.96 linear fold change corresponds to a 4% decrease in MBD. We further used multinomial logistic regression models to evaluate the associations of growth factor gene expression with categories of VPD, adjusted for confounders.
Results
The mean age of the study participants was 57.9 years (Table 1). The mean BMI was 31.3 kg/m2, which is consistent with the BMI of women attending screening mammograms at the Joanne Knight Breast Health Center. Many participants were Non-Hispanic White (62%) and African-American (35.6%). The mean VPD was 6.2% (BI-RADS category b). The mean DV and NDV were 121.2 cm3 and 2134.4 cm3, respectively.
There were positive correlations between progesterone and OPG plasma gene expression (r = 0.65, p value < 0.0001), STAT1 and STAT5 (r = 0.59, p value < 0.0001) plasma gene expression (Table 2), ESR1 and progesterone plasma gene expression (r = 0.43, p value < 0.0001) as well as prolactin and STAT5 (r = 0.43, p value < 0.0001) plasma gene expression. TNFRSF18 plasma gene expression was positively correlated with the nine other markers. RANK plasma gene expression was negatively correlated with prolactin, STAT1, and STAT5 plasma gene expression. BMI was weakly inversely correlated with gene expression of RANK pathway markers (Table 2) but not with hormone gene expression, and further adjusting the correlations for age and BMI had negligible impact on the correlation coefficients (Additional file 1: Table S1).
Table 3 shows the associations between hormone and RANK pathway plasma gene expression and VPD. Of the 10 markers evaluated ESR1, RANK, and TNFRSF18 plasma gene expression were associated with VPD. A one-unit increase in ESR1, RANK, and TNFRSF18 plasma gene expression was associated with 8% (95% CI 0–15%, p value = 0.05), 10% (95% CI 0–20%, p value = 0.04), and 4% (95% CI 0–9%, p value = 0.04) higher VPD, respectively.
We also investigated the associations of plasma gene expression across categories of VPD using multinominal logistic regression models (Table 4). The associations were similar to what we observed evaluating gene expression in the continuous form. We, however, also observed positive associations for RANKL and OPG plasma gene expression when we compared extremes of MBD. Women with extremely dense breasts (VPD > 15.5%; BI-RADS d) had a 73% (95% CI 1.05–2.85, p value = 0.03) higher plasma RANKL gene expression, and 86% (95% CI 1.10–3.14, p value = 0.02) higher plasma OPG gene expression compared with women with almost entirely fatty breasts (VPD < 3.5%; BI-RADS a). RANKL and OPG plasma gene expression was not higher among women with heterogeneously dense breasts (VPD ≥ 7.5% and < 15.5%; BI-RADS c) compared with women with almost entirely fatty breasts.
In a subset of our study participants (82 women with dense breasts) who had both circulating RANK, RANKL, and OPG and mRNA gene expression data (Additional file 1: Table S2), we observed mild positive correlations between circulating protein levels and the mRNA gene expression for RANK (r = 0.26, p value = 0.03), RANKL (r = 0.23, p value = 0.04) but not for OPG (r = − 0.03, p value = 0.81).
We further evaluated the associations of plasma gene expression with NDV and DV (Additional file 1: Table S3). A one-unit increase in plasma OPG, PGR, and TNFRSF13B gene expression was associated with 12% (95% CI 4–19%, p value = 0.003), 7% (95% CI 0–13%, p value = 0.04), and 5% lower (95% CI 1–9%, p value = 0.02) NDV, respectively. Only plasma OPG gene expression was associated with DV: a one-unit increase in OPG was associated with 8% (95% CI 0–15%, p value = 0.05) lower DV.
Discussion
To the best of our knowledge, this is the first study to investigate the associations of plasma hormone and RANK pathway gene expression with volumetric measures of MBD in postmenopausal women. We observed positive associations of ESR1, RANK, and TNFRSF18 plasma gene expression with VPD and inverse associations of PGR and OPG plasma gene expression with NDV.
VPD represents the stromal and epithelial components of fibroglandular breast tissue and is positively associated with breast cancer risk [46,47,48] while NDV represents the adipose component of breast tissue and is inversely associated with breast cancer risk in many studies [46, 49, 50]. Our finding of a positive association of plasma ESR1 gene expression with VPD is similar to that reported for circulating estradiol and percent density in some studies [11, 51, 52], while other studies have reported inverse [12, 16, 17], or no associations between circulating estradiol and percent density [13, 15, 53]. These results are difficult to directly compare as they examine plasma gene expression or circulating hormone levels. Taken together, they suggest that a singular circulating estradiol level, as determined in these studies, may not be sufficient to serve as a reliable proxy for estrogen activity.
ESR1 (ERα) regulates estrogen activity, and the ESR1 gene encodes a transcription factor with an estrogen binding domain, activating domain, and estrogen response element [54, 55]. Once estrogen binds to ESR1, proliferation is induced in both normal and neoplastic breast epithelial cells through ESR1 signaling of estrogen-responsive genes [56]. Thus, if ESR1 gene expression is increased this may lead to greater proliferation of breast tissue, culminating in greater MBD and increased breast cancer risk. This mechanism could explain the association between plasma ESR1 gene expression and VPD identified in our study.
Interestingly, a previous study using data from 79 women reported a positive association of serum estradiol level but reported an inverse association of breast tissue ESR1 gene expression with percent density in postmenopausal women [21]. The authors of this study pointed out that increased levels of estradiol have also been shown to decrease levels of ESR1 in breast cancer [57] and thus reasoned that the association between reduced ESR1 and high MBD may reflect high levels of plasma estradiol. Therefore, longitudinal studies that concomitantly explore the associations between circulating estradiol and plasma ESR1 gene expression with MBD are warranted in postmenopausal women.
The associations of serum RANK and RANKL gene expression we observed are similar to our results in premenopausal women [22]. RANK causes mammary epithelial cell proliferation perhaps via upregulation of cyclin D1 [58], and RANKL is essential for the development, formation, and differentiation of mammary glands [25, 59]. Some studies have reported associations of the RANK pathway with breast cancer pathogenesis, while others have not [60,61,62,63]. The positive association of plasma RANKL gene expression with MBD was limited to when we compared women at the extremes of MBD profiles, which suggests a nonlinear association between RANKL gene expression and MBD.
In addition, women with extremely dense breasts had higher OPG gene expression than women with almost entirely fatty breasts. OPG gene expression was inversely associated with DV and NDV. The findings were unexpected given that OPG competes with RANK for RANKL binding, thereby blocking RANK activation [64]. Hence, we hypothesized that OPG mRNA expression would be negatively associated with VPD and DV. Our findings are similar to a study that reported low OPG serum levels to be associated with high mammographic breast density (VPD and DV) [65] and contrary to another study that found no association between OPG and VPD [63]. Furthermore, some findings from preclinical studies show that OPG expression in tissue may be associated with breast tumor formation [36, 66]. Thus, the association of OPG with mammographic breast density remains unclear and deserves to be studied further. Due to the limited data on the role of OPG in breast proliferation, development, and function in humans, clinical studies are needed to elucidate the role of OPG in MBD and breast cancer development as well as how these associations may be mediated by estrogen and progesterone, given the correlations we observed across these genes.
Plasma progesterone receptor gene expression was not associated with VPD but was associated with NDV. Some studies have reported associations between circulating progesterone and percent breast density and percent dense area [13, 67, 68], while others have not [12, 15, 17]. Our finding is similar to another study that found a positive association of progesterone with absolute non-dense breast volume in premenopausal women [69]. NDV is inversely associated with breast cancer risk [46, 49, 50], suggesting that elevated progesterone gene expression may be associated with elevated breast cancer risk in postmenopausal women.
We found no association between plasma prolactin gene expression and VPD, consistent with findings from previous studies on circulating prolactin and percent dense area in postmenopausal women [17, 70], but not with others that have reported positive associations [12, 71]. One study used an immunoassay rather than circulating hormones to quantify prolactin levels and determined that postmenopausal women with high prolactin immunoassay profiles had higher breast dense area and lower non-dense area than those with lower prolactin immunoassay readings [72].
We observed correlations between plasma RANK pathway gene expression and hormone gene expression, which is an indication of the crosstalk between these markers and may provide further biological insights into the complex pathways through which the markers influence MBD and breast cancer risk. Progesterone upregulates RANKL expression [24, 25] and interacts with the RANK/RANKL pathway through signal transducer and activator of transcription (STAT) signaling [26, 27], while OPG mRNA transcription in healthy breast tissue is regulated by estrogen [73, 74]. Further clinical studies are needed to characterize the interrelationships between these markers and how they influence MBD and breast cancer development. The inverse correlations of BMI with OPG gene expression we observed are similar to what has been reported for their circulating levels [75] but we did not observe positive correlations of BMI with ESR1 and PRL gene expression, in contrast to what has been reported for their circulating levels [76]. Other studies evaluating correlations of BMI with hormone gene expression are needed.
Our study has several strengths. Study participants were recruited among women attending annual routine screening mammograms. We analyzed plasma hormone gene expression rather than circulating hormone levels, and we did compare the gene expression to protein levels in a subset of participants. However, we did not compare the mRNA gene expression in the plasma to the gene expression in the breast tissue since study participants were cancer-free women recruited during their annual screening mammogram. Future studies integrating plasma gene expression, target tissue gene expression, and circulating protein levels as biomarkers in elucidating MBD for breast cancer risk are encouraged.
One limitation of our study is that the sample size was not large enough to perform mediation analyses between hormone and RANK gene expression on MBD or to conduct analyses stratified by BMI and race. We did not profile plasma gene expression of other hormones such as androgens and sex hormone-binding globulin, and parathyroid hormone, a known regulator of the RANK pathway. Future studies evaluating how these hormones are associated with MBD will be needed.
Conclusions
In conclusion, we observed positive associations of ESR1, RANK, and TNFRSF18 plasma gene expression with VPD in postmenopausal women. Women with extremely dense breasts had higher RANKL and OPG plasma gene expression than women with entirely fatty breasts. These findings require validation within other study populations.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional file 1.
Abbreviations
- MBD:
-
Mammographic breast density
- RANK:
-
Receptor activator of nuclear factor-kB
- RANKL:
-
Receptor activator of nuclear factor kappa-Β ligand
- VPD:
-
Volumetric percent density
- DV:
-
Dense volume
- NDV:
-
Non-dense volume
- PRL:
-
Prolactin
- PGR:
-
Progesterone receptor
- ESR1:
-
Estrogen receptor 1
- STAT1:
-
Signal transducer and activator of transcription 1
- STAT5:
-
Signal transducer and activator of transcription 5
- TNFRSF13B:
-
Tumor necrosis factor receptor superfamily member 13B
- TNFRSF18:
-
Tumor necrosis factor receptor superfamily member 18
- OPG:
-
Osteoprotegerin
References
Boyd NF, Guo H, Martin LJ, Sun L, Stone J, Fishell E, et al. Mammographic density and the risk and detection of breast cancer. N Engl J Med. 2007;356(3):227–36.
Boyd NF, Rommens JM, Vogt K, Lee V, Hopper JL, Yaffe MJ, et al. Mammographic breast density as an intermediate phenotype for breast cancer. Lancet Oncol. 2005;6(10):798–808.
Cuzick J, Warwick J, Pinney E, Duffy SW, Cawthorn S, Howell A, et al. Tamoxifen-induced reduction in mammographic density and breast cancer risk reduction: a nested case-control study. J Natl Cancer Inst. 2011;103(9):744–52.
Leehy KA, Truong TH, Mauro LJ, Lange CA. Progesterone receptors (PR) mediate STAT actions: PR and prolactin receptor signaling crosstalk in breast cancer models. J Steroid Biochem Mol Biol. 2018;176:88–93.
McTiernan A, Martin CF, Peck JD, Aragaki AK, Chlebowski RT, Pisano ED, et al. Estrogen-plus-progestin use and mammographic density in postmenopausal women: Women’s Health Initiative randomized trial. J Natl Cancer Inst. 2005;97(18):1366–76.
Marchesoni D, Driul L, Ianni A, Fabiani G, Della Martina M, Zuiani C, et al. Postmenopausal hormone therapy and mammographic breast density. Maturitas. 2006;53(1):59–64.
Persson I, Thurfjell E, Holmberg L. Effect of estrogen and estrogen-progestin replacement regimens on mammographic breast parenchymal density. J Clin Oncol Off J Am Soc Clin Oncol. 1997;15(10):3201–7.
Rutter CM, Mandelson MT, Laya MB, Seger DJ, Taplin S. Changes in breast density associated with initiation, discontinuation, and continuing use of hormone replacement therapy. JAMA. 2001;285(2):171–6.
Buist DSM, Anderson ML, Reed SD, Aiello Bowles EJ, Fitzgibbons ED, Gandara JC, et al. Short-term hormone therapy suspension and mammography recall: a randomized trial. Ann Intern Med. 2009;150(11):752–65.
Becker S, Kaaks R. Exogenous and endogenous hormones, mammographic density and breast cancer risk: Can mammographic density be considered an intermediate marker of risk? Recent Results Cancer Res. 2009;181:135–57.
Greendale GA, Palla SL, Ursin G, Laughlin GA, Crandall C, Pike MC, et al. The association of endogenous sex steroids and sex steroid binding proteins with mammographic density: results from the postmenopausal estrogen/progestin interventions mammographic density study. Am J Epidemiol. 2005;162(9):826–34.
Boyd NF, Stone J, Martin LJ, Jong R, Fishell E, Yaffe M, et al. The association of breast mitogens with mammographic densities. Br J Cancer. 2002;87(8):876–82.
Sprague BL, Trentham-Dietz A, Gangnon RE, Buist DS, Burnside ES, Bowles EJ, et al. Circulating sex hormones and mammographic breast density among postmenopausal women. Horm Cancer. 2011;2(1):62–72.
Tamimi RM, Byrne C, Colditz GA, Hankinson SE. Endogenous hormone levels, mammographic density, and subsequent risk of breast cancer in postmenopausal women. JNCI J Natl Cancer Inst. 2007;99(15):1178–87.
Warren R, Skinner J, Sala E, Denton E, Dowsett M, Folkerd E, et al. Associations among mammographic density, circulating sex hormones, and polymorphisms in sex hormone metabolism genes in postmenopausal women. Cancer Epidemiol Biomarkers Prev. 2006;15(8):1502–8.
Aiello EJ, Tworoger SS, Yasui Y, Stanczyk FZ, Potter J, Ulrich CM, et al. Associations among circulating sex hormones, insulin-like growth factor, lipids, and mammographic density in postmenopausal women. Cancer Epidemiol Biomark Prev. 2005;14(6):1411–7.
Tamimi RM, Hankinson SE, Colditz GA, Byrne C. Endogenous sex hormone levels and mammographic density among postmenopausal women. Cancer Epidemiol Biomark Prev. 2005;14(11):2641–7.
Sun X, Gierach GL, Sandhu R, Williams T, Midkiff BR, Lissowska J, et al. Relationship of mammographic density and gene expression: analysis of normal breast tissue surrounding breast cancer. Clin Cancer Res. 2013;19(18):4972–82.
Haakensen VD, Biong M, Lingjaerde OC, Holmen MM, Frantzen JO, Chen Y, et al. Expression levels of uridine 5’-diphospho-glucuronosyltransferase genes in breast tissue from healthy women are associated with mammographic density. Breast Cancer Res. 2010;12(4):R65.
Yang WT, Lewis MT, Hess K, Wong H, Tsimelzon A, Karadag N, et al. Decreased TGFbeta signaling and increased COX2 expression in high risk women with increased mammographic breast density. Breast Cancer Res Treat. 2010;119(2):305–14.
Haakensen VD, Biong M, Lingjærde OC, Holmen MM, Frantzen JO, Chen Y, et al. Expression levels of uridine 5’-diphospho-glucuronosyltransferase genes in breast tissue from healthy women are associated with mammographic density. Breast Cancer Res. 2010;12(4):R65.
Toriola AT, Dang HX, Hagemann IS, Appleton CM, Colditz GA, Luo J, et al. Increased breast tissue receptor activator of nuclear factor-κB ligand (RANKL) gene expression is associated with higher mammographic density in premenopausal women. Oncotarget. 2017;8(43):73787–92.
Fernandez-Valdivia R, Lydon JP. From the ranks of mammary progesterone mediators, RANKL takes the spotlight. Mol Cell Endocrinol. 2012;357(1–2):91–100.
Dougall WC. Molecular pathways: osteoclast-dependent and osteoclast-independent roles of the RANKL/RANK/OPG pathway in tumorigenesis and metastasis. Clin Cancer Res. 2012;18(2):326.
Fata JE, Kong YY, Li J, Sasaki T, Irie-Sasaki J, Moorehead RA, et al. The osteoclast differentiation factor osteoprotegerin-ligand is essential for mammary gland development. Cell. 2000;103(1):41–50.
Srivastava S, Matsuda M, Hou Z, Bailey JP, Kitazawa R, Herbst MP, et al. Receptor activator of NF-κB ligand induction via Jak2 and Stat5a in mammary epithelial cells. J Biol Chem. 2003;278(46):46171–8.
Obr AE, Grimm SL, Bishop KA, Pike JW, Lydon JP, Edwards DP. Progesterone receptor and Stat5 signaling cross talk through RANKL in mammary epithelial cells. Mol Endocrinol. 2013;27(11):1808–24.
Denlinger CS, Sanft T, Baker KS, Baxi S, Broderick G, Demark-Wahnefried W, et al. Survivorship, Version 2.2017, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2017;15(9):1140–63.
Alimujiang A, Imm KR, Appleton CM, Colditz GA, Berkey CS, Toriola AT. Adiposity at age 10 and mammographic density among premenopausal women. Cancer Prev Res (Phila). 2018;11(5):287–94.
Lee HN, Sohn YM, Han KH. Comparison of mammographic density estimation by Volpara software with radiologists’ visual assessment: analysis of clinical-radiologic factors affecting discrepancy between them. Acta Radiol. 2015;56(9):1061–8.
Ko SY, Kim EK, Kim MJ, Moon HJ. Mammographic density estimation with automated volumetric breast density measurement. Korean J Radiol. 2014;15(3):313–21.
Gry M, Rimini R, Stromberg S, Asplund A, Ponten F, Uhlen M, et al. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genomics. 2009;10(1):365.
Shankavaram UT, Reinhold WC, Nishizuka S, Major S, Morita D, Chary KK, et al. Transcript and protein expression profiles of the NCI-60 cancer cell panel: an integromic microarray study. Mol Cancer Ther. 2007;6(3):820–32.
de Sousa Abreu R, Penalva LO, Marcotte EM, Vogel C. Global signatures of protein and mRNA expression levels. Mol Biosyst. 2009;5(12):1512–26.
Ming J, Cronin SJF, Penninger JM. Targeting the RANKL/RANK/OPG axis for cancer therapy. Front Oncol. 2020. https://doi.org/10.3389/fonc.2020.01283.
Infante M, Fabi A, Cognetti F, Gorini S, Caprio M, Fabbri A. RANKL/RANK/OPG system beyond bone remodeling: involvement in breast cancer and clinical perspectives. J Exp Clin Cancer Res. 2019;38(1):12.
Sigl V, Jones LP, Penninger JM. RANKL/RANK: from bone loss to the prevention of breast cancer. Open Biol. 2016;6(11):160230.
Cordero A, Pellegrini P, Sanz-Moreno A, Trinidad EM, Serra-Musach J, Deshpande C, et al. Rankl impairs lactogenic differentiation through inhibition of the prolactin/Stat5 pathway at midgestation. Stem Cells. 2016;34(4):1027–39.
Furth PA, Nakles RE, Millman S, Diaz-Cruz ES, Cabrera MC. Signal transducer and activator of transcription 5 as a key signaling pathway in normal mammary gland developmental biology and breast cancer. Breast Cancer Res. 2011;13(5):220.
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 2008;26(3):317–25.
Kojima K, April C, Canasto-Chibuque C, Chen X, Deshmukh M, Venkatesh A, et al. Transcriptome profiling of archived sectioned formalin-fixed paraffin-embedded (AS-FFPE) tissue for disease classification. PLoS ONE. 2014;9(1):e86961.
Payton JE, Grieselhuber NR, Chang L-W, Murakami M, Geiss GK, Link DC, et al. High throughput digital quantification of mRNA abundance in primary human acute myeloid leukemia samples. J Clin Investig. 2009;119(6):1714–26.
Reis PP, Waldron L, Goswami RS, Xu W, Xuan Y, Perez-Ordonez B, et al. mRNA transcript quantification in archival samples using multiplexed, color-coded probes. BMC Biotechnol. 2011;11(1):46.
Speranza E, Altamura LA, Kulcsar K, Bixler SL, Rossi CA, Schoepp RJ, et al. Comparison of transcriptomic platforms for analysis of whole blood from Ebola-infected cynomolgus macaques. Sci Rep. 2017;7(1):14756.
Bracht JWP, Gimenez-Capitan A, Huang C-Y, Potie N, Pedraz-Valdunciel C, Warren S, et al. Analysis of extracellular vesicle mRNA derived from plasma using the nCounter platform. Sci Rep. 2021;11(1):3712.
Pettersson A, Graff RE, Ursin G, Santos Silva ID, McCormack V, Baglietto L, et al. Mammographic density phenotypes and risk of breast cancer: a meta-analysis. J Natl Cancer Inst. 2014;106(5):1–11.
Stone J, Ding J, Warren RM, Duffy SW, Hopper JL. Using mammographic density to predict breast cancer risk: dense area or percentage dense area. Breast Cancer Res BCR. 2010;12(6):R97-R.
Vachon CM, Brandt KR, Ghosh K, Scott CG, Maloney SD, Carston MJ, et al. Mammographic breast density as a general marker of breast cancer risk. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2007;16(1):43–9.
Pettersson A, Hankinson SE, Willett WC, Lagiou P, Trichopoulos D, Tamimi RM. Nondense mammographic area and risk of breast cancer. Breast Cancer Res. 2011;13(5):R100.
Yaghjyan L, Colditz GA, Rosner B, Tamimi RM. Mammographic breast density and breast cancer risk: interactions of percent density, absolute dense, and non-dense areas with breast cancer risk factors. Breast Cancer Res Treat. 2015;150(1):181–9.
Johansson H, Gandini S, Bonanni B, Mariette F, Guerrieri-Gonzaga A, Serrano D, et al. Relationships between circulating hormone levels, mammographic percent density and breast cancer risk factors in postmenopausal women. Breast Cancer Res Treat. 2008;108(1):57–67.
Haakensen VD, Bjøro T, Lüders T, Riis M, Bukholm IK, Kristensen VN, et al. Serum estradiol levels associated with specific gene expression patterns in normal breast tissue and in breast carcinomas. BMC Cancer. 2011;11(1):332.
Tamimi RM, Hankinson SE, Colditz GA, Byrne C. Endogenous sex hormone levels and mammographic density among postmenopausal women. Cancer Epidemiol Biomark Prev. 2005;14(11 Pt 1):2641–7.
Yaghjyan L, Colditz GA, Rosner B, Tamimi RM. Mammographic breast density and subsequent risk of breast cancer in postmenopausal women according to the time since the mammogram. Cancer Epidemiol Biomark Prev Publ Am Assoc Cancer Res Cosponsored Am Soc Prev Oncol. 2013;22(6):1110–7.
Zhang Y, Zhang M, Yuan X, Zhang Z, Zhang P, Chao H, et al. Association between ESR1 PvuII, XbaI, and P325P polymorphisms and breast cancer susceptibility: a meta-analysis. Med Sci Monit. 2015;21:2986–96.
Dumas I, Diorio C. Estrogen pathway polymorphisms and mammographic density. Anticancer Res. 2011;31(12):4369–86.
Dunbier AK, Anderson H, Ghazoui Z, Folkerd EJ, A’Hern R, Crowder RJ, et al. Relationship between plasma estradiol levels and estrogen-responsive gene expression in estrogen receptor–positive breast cancer in postmenopausal women. J Clin Oncol. 2010;28(7):1161–7.
Gonzalez-Suarez E, Jacob AP, Jones J, Miller R, Roudier-Meyer MP, Erwert R, et al. RANK ligand mediates progestin-induced mammary epithelial proliferation and carcinogenesis. Nature. 2010;468(7320):103–7.
Fernandez-Valdivia R, Mukherjee A, Ying Y, Li J, Paquet M, DeMayo FJ, et al. The RANKL signaling axis is sufficient to elicit ductal side-branching and alveologenesis in the mammary gland of the virgin mouse. Dev Biol. 2009;328(1):127–39.
Odén L, Akbari M, Zaman T, Singer CF, Sun P, Narod SA, et al. Plasma osteoprotegerin and breast cancer risk in BRCA1 and BRCA2 mutation carriers. Oncotarget. 2016;7(52):86687–94.
Kiechl S, Schramek D, Widschwendter M, Fourkala EO, Zaikin A, Jones A, et al. Aberrant regulation of RANKL/OPG in women at high risk of developing breast cancer. Oncotarget. 2017;8(3):3811–25.
Sarink D, Schock H, Johnson T, Overvad K, Holm M, Tjønneland A, et al. Circulating RANKL and RANKL/OPG and breast cancer risk by ER and PR subtype: results from the EPIC cohort. Cancer Prev Res (Phila). 2017;10(9):525–34.
Kotsopoulos J, McGee EE, Lozano-Esparza S, Garber JE, Ligibel J, Collins LC, et al. Premenopausal plasma osteoprotegerin and breast cancer risk: a case-control analysis nested within the nurses’ health study II. Cancer Epidemiol Biomark Prev. 2020;29(6):1264–70.
Hanada R, Hanada T, Penninger JM. Physiology and pathophysiology of the RANKL/RANK system. Biol Chem. 2010;391(12):1365–70.
Moran O, Zaman T, Eisen A, Demsky R, Blackmore K, Knight JA, et al. Serum osteoprotegerin levels and mammographic density among high-risk women. Cancer Causes Control. 2018;29(6):507–17.
Weichhaus M, Chung STM, Connelly L. Osteoprotegerin in breast cancer: beyond bone remodeling. Mol Cancer. 2015;14:117.
Gabrielson M, Azam S, Hardell E, Holm M, Ubhayasekera KA, Eriksson M, et al. Hormonal determinants of mammographic density and density change. Breast Cancer Res. 2020;22(1):95.
Hada M, Oh H, Fan S, Falk RT, Geller B, Vacek P, et al. Relationship of serum progesterone and progesterone metabolites with mammographic breast density and terminal ductal lobular unit involution among women undergoing diagnostic breast biopsy. J Clin Med. 2020;9(1):245.
Jung S, Stanczyk FZ, Egleston BL, Snetselaar LG, Stevens VJ, Shepherd JA, et al. Endogenous sex hormones and breast density in young women. Cancer Epidemiol Biomark Prev. 2015;24(2):369–78.
Maskarinec G, Takata Y, Chen Z, Gram IT, Nagata C, Pagano I, et al. IGF-I and mammographic density in four geographic locations: a pooled analysis. Int J Cancer. 2007;121(8):1786–92.
Greendale GA, Huang MH, Ursin G, Ingles S, Stanczyk F, Crandall C, et al. Serum prolactin levels are positively associated with mammographic density in postmenopausal women. Breast Cancer Res Treat. 2007;105(3):337–46.
Rice MS, Tworoger SS, Bertrand KA, Hankinson SE, Rosner BA, Feeney YB, et al. Immunoassay and Nb2 lymphoma bioassay prolactin levels and mammographic density in premenopausal and postmenopausal women the Nurses’ Health Studies. Breast Cancer Res Treat. 2015;149(1):245–53.
Hofbauer LC, Khosla S, Dunstan CR, Lacey DL, Spelsberg TC, Riggs BL. Estrogen stimulates gene expression and protein production of osteoprotegerin in human osteoblastic cells. Endocrinology. 1999;140(9):4367–70.
Saika M, Inoue D, Kido S, Matsumoto T. 17beta-estradiol stimulates expression of osteoprotegerin by a mouse stromal cell line, ST-2, via estrogen receptor-alpha. Endocrinology. 2001;142(6):2205–12.
Ashley DT, O’Sullivan EP, Davenport C, Devlin N, Crowley RK, McCaffrey N, et al. Similar to adiponectin, serum levels of osteoprotegerin are associated with obesity in healthy subjects. Metabolism. 2011;60(7):994–1000.
McTiernan A, Wu L, Chen C, Chlebowski R, Mossavar-Rahmani Y, Modugno F, et al. Relation of BMI and physical activity to sex hormones in postmenopausal women. Obesity (Silver Spring). 2006;14(9):1662–77.
Acknowledgements
We would like to thank all of the women who participated in this study.
Funding
This work was supported by funding from NIH/NCI (R21CA216515, R01CA246592, and R37CA235602 to Adetunji T. Toriola).
Author information
Authors and Affiliations
Contributions
ATT designed the study. ATT acquired the data. MW and SX analyzed the data. RM, SX, GC, and ATT interpreted the data. RM wrote the manuscript drafts. RM, MW, SX, CM, GC, and ATT revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This work was approved by Washington University’s ethics committees. All participants consented to participate in this work.
Consent for publication
Not applicable.
Competing interests
The authors have no potential competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Table S1: Correlations between Hormones and RANK Pathway Gene Expression in Postmenopausal Women, adjusted for age and BMI. Table S2: Correlations between Circulating RANK, sRANKL, OPG Protein and Their Gene Expression Levels Among 82 Women with Dense Breasts. Table S3: Associations of Hormone, RANK Pathway Gene Expression with Non-Dense Volume and Dense Volume.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Mintz, R., Wang, M., Xu, S. et al. Hormone and receptor activator of NF-κB (RANK) pathway gene expression in plasma and mammographic breast density in postmenopausal women. Breast Cancer Res 24, 28 (2022). https://doi.org/10.1186/s13058-022-01522-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13058-022-01522-2