Abstract
Background
Chinese medicine Xuebijing (XBJ) has proven to be effective in the treatment of mild coronavirus disease 2019 (COVID-19) cases. But the bioactive compounds and potential mechanisms of XBJ for COVID-19 prevention and treatment are unclear. This study aimed to examine the potential effector mechanisms of XBJ on COVID-19 based on network pharmacology.
Methods
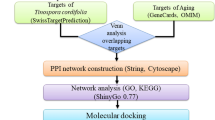
We searched Chinese and international papers to obtain the active ingredients of XBJ. Then, we compiled COVID-19 disease targets from the GeneCards gene database and via literature searches. Next, we used the SwissTargetPrediction database to predict XBJ’s effector targets and map them to the abovementioned COVID-19 disease targets in order to obtain potential therapeutic targets of XBJ. Cytoscape software version 3.7.0 was used to construct a “XBJ active-compound-potential-effector target” network and protein-protein interaction (PPI) network, and then to carry out network topology analysis of potential targets. We used the ClueGO and CluePedia plugins in Cytoscape to conduct gene ontology (GO) biological process (BP) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathway enrichment analysis of XBJ’s effector targets. We used AutoDock vina and PyMOL software for molecular docking.
Results
We obtained 144 potential COVID-19 effector targets of XBJ. Fourteen of these targets-glyceraldehyde 3-phosphate dehydrogenase (GAPDH), albumin (ALB), tumor necrosis factor (TNF), epidermal growth factor receptor (EGFR), mitogen-activated protein kinase 1 (MAPK1), Caspase-3 (CASP3), signal transducer and activator of transcription 3 (STAT3), MAPK8, prostaglandin-endoperoxide synthase 2 (PTGS2), JUN, interleukin-2 (IL-2), estrogen receptor 1 (ESR1), and MAPK14 had degree values > 40 and therefore could be considered key targets. They participated in extracellular signal–regulated kinase 1 and 2 (ERK1, ERK2) cascade, the T-cell receptor signaling pathway, activation of MAPK activity, cellular response to lipopolysaccharide, and other inflammation- and immune-related BPs. XBJ exerted its therapeutic effects through the renin-angiotensin system (RAS), nuclear factor κ-light-chain-enhancer of activated B cells (NF-κB), MAPK, phosphatidylinositol-4, 5-bisphosphate 3-kinase (PI3K)-protein kinase B (Akt)-vascular endothelial growth factor (VEGF), toll-like receptor (TLR), TNF, and inflammatory-mediator regulation of transient receptor potential (TRP) signaling pathways to ultimately construct a “drug-ingredient-target-pathway” effector network. The molecular docking results showed that the core 18 effective ingredients had a docking score of less than − 4.0 with those top 10 targets.
Conclusion
The active ingredients of XBJ regulated different genes, acted on different pathways, and synergistically produced anti-inflammatory and immune-regulatory effects, which fully demonstrated the synergistic effects of different components on multiple targets and pathways. Our study demonstrated that key ingredients and their targets have potential binding activity, the existing studies on the pharmacological mechanisms of XBJ in the treatment of sepsis and severe pneumonia, could explain the effector mechanism of XBJ in COVID-19 treatment, and those provided a preliminary examination of the potential effector mechanism in this disease.
Similar content being viewed by others
Introduction
In December 2019, the Huanan Seafood Wholesale Market in Wuhan, Hubei Province, China, became the epicenter of an outbreak of a pneumonia of unknown etiology, which attracted a great deal of attention in China and the rest of the world. Chinese scientists quickly isolated a novel coronavirus from patients, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19) [1]. As of March 29, 2020, the global number of confirmed cases was 634,835 and the global death toll was 29,891. Currently, scientists around the world are conducting an exhaustive search for effective antiviral drugs. However, the only feasible method at this writing is the use of various broad-spectrum antivirals, such as nucleoside analogs and human immunodeficiency virus (HIV) protease inhibitors [2]. Up to now, no COVID-19 specific antiviral drugs or vaccines have been developed, and the aforementioned drugs can only reduce viral infection [3].
Many results from clinical practice show that TCM plays an important role in COVID-19 treatment and has brought new hope for controlling this disease [4]. Xuebijing (XBJ) was included in the Diagnosis and Treatment Plan for Coronavirus Disease 2019 (interim 7th edition) that was jointly released by the National Health Commission and National Administration of Traditional Chinese Medicine. XBJ is composed of extracts of Carthamus tinctorius, Paeonia anomala, Ligusticum striatum, Salvia miltiorrhiza, and Angelica sinensis [5]. This compound can boost circulation, relieve stasis, and clear blocked meridians, and it is widely used in China to treat active inflammation [6]. In 2004, XBJ was approved by the National Medical Products Administration for the treatment of systemic inflammatory-response syndrome, sepsis, and multiple-organ dysfunction syndrome (MODS) [7, 8]. Studies show that XBJ treatment can reduce the secretion of pro-inflammatory cytokines such as interleukin (IL)-6, IL-13, and tumor necrosis factor alpha (TNF-α) to alleviate inflammation and thereby inhibit liver damage [9]. Chen et al. showed that XBJ treatment can decrease oxidative stress (OS) and levels of pro-inflammatory cytokines [10]. Li et al. showed that XBJ can regulate immune response, including reducing inflammatory mediators and bacterial load, and plays a protective role in bacterial infections, particularly those caused by drug-resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA) [11]. A real-world study also pointed out that the incidence of adverse reactions from XBJ in clinical practice is low (0.3%), and most adverse reactions are mild. Therefore, XBJ could be a safe and effective drug for treating COVID-19, but its specific molecular mechanisms are still unknown.
Network pharmacology (NP) uses drug, compound, gene, and disease database information to construct drug-target, target-disease, and drug-disease interaction networks in order to reveal the complex mechanisms of TCM formulations that have multiple targets and multiple component characteristics [12]. The concepts of NP share many similarities with the holistic view of TCM, as both use systemic methods to treat complex diseases such as cancer. This provides a basis for the transition from empirical medicine to evidence-based medicine [13].
Therefore, we employ NP to initially explore the potential molecular mechanism of XBJ on COVID-19. The basis for our NP study rests on the notions that the proteins that associate with and functionally govern viral infection are localized in the corresponding subnetwork within the comprehensive human interactome network. The latest research shows that host cell pathways regulated by SARS-CoV-2 infection and showed that inhibiting these pathways can prevent virus replication in human cells [14]. A drug with multiple targets, such as XBJ, if it is to be effective against an HCoV, its direct or indirect target proteins should be within or related to the corresponding subnetwork in the human protein-protein interactome.
In terms of potential therapeutic targets, research shows that the 2019-nCoV/SARS-CoV-2 shares the highest nucleotide sequence identity (79.7%) with SARS-CoV among the six other known pathogenic HCoVs [15]. Full-length genome sequences from five patients at an early stage of COVID-19 outbreak showed that the sequences are almost identical and share a 79.6% sequence identity to SARS-CoV, what’s more, 2019-nCoV is 96% identical at the whole-genome level to a bat coronavirus [16], and the amino acid sequences and predicted protein structures of the receptor-binding domain (RBD) of SARS-CoV2 and SARS-CoV share a high similarities [17], all of these indicate that the host proteins of SARS-CoV were considered potential therapeutic targets for COVID-19.
In addition, viruses including coronaviruses like COVID-19 require host cellular factors for successful replication during infection. Study on virus-host protein-protein interactions (PPIs) provides an effective method toward elucidating the mechanisms of viral infection [18, 19], so the virus-host interactome may offer a strategy for the treatment of viral infections, and the human pathogenic coronaviruses SARS-CoV2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV) belong to the family Coronaviridae and the genus Betacoronavirus, with similar infectivity, pathogenicity, and related clinical characteristics. Therefore, in our study, the host proteins of the relevant coronaviruses are considered as potential therapeutic targets for COVID-19. As well as angiotensin-converting enzyme 2 (ACE2) and coronavirus pneumonia related targets.
It is essential to integrate drug-target networks, HCoV-host interactions and human protein-protein interactome network. Based on chemical-matteromics study results for XBJ, we employed NP to construct a “component-target-pathway” network model in order to comprehensively and systematically predict potential effector targets and pathways of this compound’s main chemical components in COVID-19 treatment. These findings will provide a scientific basis for further research into effective substances and mechanisms in XBJ treatment of COVID-19, the overall workflow of this study was presented in Fig. 1.
Materials and methods
Collection of potential active ingredients of XBJ
We searched the global scientific literature to determine the active ingredients of XBJ. The PubChem (https://pubchem.ncbi.nlm.nih.gov/, National Center for Biotechnology Information, 8600 Rockville Pike, Bethesda, MD, USA) was used for cross-validation and to acquire the molecular structures of potential active chemical components, which we stored in canonical simplified molecular-input line-entry system (SMILES) format.
Prediction of potential effector targets of XBJ
To predict potential effector targets, we used the SwissTargetPrediction database [20] (STP, http://www.swisstargetprediction.ch, version 2019, designed by Swiss Institute of Bioinformatics, Quartier Sorge - Batiment Amphipole 1015 Lausanne, Switzerland). We input the aforementioned potential active ingredients in SMILES format into this database, with “humans” (Homo sapiens) as the study species, to obtain the potential effector targets of the component compounds; results were stored in csv format. After compilation and removal of repetitions, we obtained the potential effector targets of XBJ.
Screening of potential therapeutic targets of COVID-19
Three sources were used to obtain potential therapeutic targets of COVID-19. First, we obtained the COVID-19 disease target set by searching the GeneCards gene database (http://www.genecards.org, designed by The Weizmann Institute of Science, 234 Herzl Street, POB 26, Rehovot, Israel) [21] on the phrase “coronavirus pneumonia”. Second, we searched the literature to collect potential therapeutic targets of COVID-19. ACE is reported to be the receptor for SARS-CoV [22] and is also believed to be that for SARS-CoV-2. We used single-cell sequencing results for colon epithelial cells [23] to extract genes that are co-expressed with ACE2, which we matched with human targets as potential therapeutic targets for COVID-19. Third, we downloaded human coronavirus (HCoV)-related host proteins from the appendices of one study [15]. Specifically, these host proteins are either the direct targets of HCoV proteins or are involved in crucial pathways of HCoV infection. By regulating the relevant coronavirus host protein could lead to a therapeutic regimen to treat COVID-19 [24].The relevant coronaviruses include SARS-CoV, MERS-CoV, infectious-bronchitis virus (IBV), mouse hepatitis virus (MHV), HCoV-229E, and HCoV-NL63; the host proteins were considered potential therapeutic targets for COVID-19. Finally, we obtained intersections between the active-ingredient-related targets and the disease-related targets from the three abovementioned sources. Intersectional targets were considered potential therapeutic targets of XBJ in COVID-19.
Construction of a network of active ingredients and effector targets
We input the abovementioned potential active compounds of XBJ and their potential effector targets into Cytoscape software (http://www.cytoscape.org, Version 3.7.0, designed by Department of Bioengineering, University of California-San Diego, California, USA) [25] to plot a “XBJ active-compound-potential effector target” network analysis map. On this map, different nodes represented potential active compounds and effector targets of XBJ, and the map’s edges showed relationships between these two factors.
Protein-protein interaction (PPI) analysis and network topology analysis
We input the potential therapeutic targets of XBJ into the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING, https://string-db.org/, supported by ELIXIR, Wellcome Genome Campus, Hinxton, Cambridgeshire, UK) [26] and selected humans as the species to obtain PPI data. Next, we input this data into Cytoscape to plot a PPI network map. The cytoHubba (Predicts and explores important nodes and subnetworks in a given network by several topological algorithms, provided by Institute of Information Science, Academia Sinica, Taiwan) [27] plugin in Cytoscape was used for network topology analysis, and results were sorted by the degree value.
Gene functional annotation of potential effector targets
The org.Hs.eg.db (https://www.bioconductor.org/, a package for the genome wide annotation of Human, primarily based on mapping using Entrez Gene identifiers, version 3.1.1) and the clusterProfiler (https://www.bioconductor.org, a package implements methods to analyze and visualize functional profiles of gene and gene clusters, version 3.1.1) [28] on the R 3.5.2, were used to perform the gene ontology (GO) biological process (BP) enrichment analysis [29] and the Kyoto Gene and Genome Encyclopedia (KEGG) enrichment analysis [30] of the potential effector targets, and P < 0.05 were selected. We used the ClueGO (version 2.5.4, creates and visualizes a functionally grouped network of terms/pathways) [31] and CluePedia (version 1.5.4, a ClueGO plugin for pathway insights using integrated experimental and in silico data) (both provided by Laboratory of Integrative Cancer Immunology, Paris, France) [32] plugins in Cytoscape, to perform WikiPathways enrichment analysis of the potential effector targets. We chose the analysis mode of Functional analysis, load marker list is Homo sapiens (9606) and Symbol ID was input, then we selected “WikiPathways-503 terms/pathways with 6558 available unique genes-27.02.2019”. Regarding statistical options, we employed two-sided hypergeometric test and bonferroni step down for p value correction, and P < 0.01 were selected.
Analysis of the binding activity of ingredients with its targets by molecular docking
We performed molecular docking by using Auto Dock vina [33] (version 1.1.2, an open-source program for doing molecular docking designed and implemented by Dr. Oleg Trott in the Molecular Graphics Lab at The Scripps Research Institute) to verify the binding activity of the core ingredients of XBJ with its potential targets of COVID-19. The 3D structure of the ingredient was obtained in the PubChem and the core target protein in the Protein Data Bank (https://www.rcsb.org/). The screening criteria for target proteins were: (1) the organisms were Homo sapiens; (2) the analysis method was X-ray diffraction; (3) resolution < 2.5A; (4) the released date was as late as possible; (5) preference was given to those containing unique ligands. The ingredients and protein structures were processed by AutoDockTools (version 1.5.6). The combinations of best docking score were visualized by PyMOL.
Results
Network analysis of active ingredients and effector targets
We selected 30 active ingredients of XBJ that had been detected by liquid chromatography-mass spectrometry (LC-MS) [34]. Results are shown in Table 1. 451 targets of XBJ for Homo sapiens were obtained based on Swiss database (The details of target information are shown in the Supplementary file, Table S1; 451 targets of XBJ are shown in Supplementary file, Table S2). We obtained 251 potential therapeutic targets of COVID-19 from the GeneCards database (Supplementary file, Table S3), 119 therapeutic targets were obtained from the systemic literature search (Supplementary file, Table S4) and Single-cell sequencing was used to obtain 3525 gene targets that were co-expressed with ACE (Supplementary file, Table S5). We obtained 144 potential COVID-19 therapeutic targets of XBJ in total after deduplication (Fig. 2 and Supplementary file, Table S6). We used Cytoscape to plot a “component-target” network relationship map (Fig. 3). Four ingredients had no targets that overlapped with potential therapeutic targets for COVID-19, so this network included 170 nodes, which were comprised of 26 XBJ components matched to 144 disease-related targets.
Network analysis of potential therapeutic targets
We input the PPI information obtained from STRING into Cytoscape to plot the PPI network, and then we used cytoHubba for network topology analysis. Table 2 shows the results, Fig. 4 shows the visualization results, and both show the top 50 potential therapeutic targets by degree values. The details of network topology analysis results of 114 potential therapeutic targets are shown in the Supplementary file, Table S7. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), TNF, mitogen-activated protein kinase 3 (MAPK3), Caspase-3 (CASP3), epidermal growth factor receptor (EGFR), MAPK1, prostaglandin-endoperoxide synthase 2 (PTGS2), signal transducer and activator of transcription 3 (STAT3), and MAPK8 all had degree values > 40, showing that these proteins occupied important positions in the PPI network.
GO gene function and KEGG signaling pathway enrichment analyses
GO BP enrichment analysis showed that the potential therapeutic targets of XBJ involved 228 BPs. From those GO terms, it suggests that XBJ compounds may regulate the inflammatory response, vasoconstriction, and cytosolic calcium ion concentration; response to the corticosteroid, reactive oxygen species, oxidative stress, lipopolysaccharide, and biotic stimulus, and involve in cell chemotaxis, peptidyl-serine modification, leukocyte migration. This indicated that the active ingredients of XBJ could exert their effects through multiple BPs (Fig. 5 and Supplementary file, Table S8).
KEGG signaling pathway enrichment analysis showed that XBJ acted on COVID-19 through 94 pathways. From those KEGG terms, it shows that targets were significantly enriched in multiple pathways such as IL-17, B cell receptor, PI3K − Akt, VEGF, T cell receptor, NF-kappa B, TNF, HIF-1 signaling pathway, and vascular smooth muscle contraction, platelet activation, arachidonic acid metabolism, apoptosis. (Fig. 6 and Supplementary file, Table S9).
WikiPathways enrichment analysis showed that XBJ acted on COVID-19 mainly through 40 pathways, such as HIF-1 survival, TWEAK, corticotropin-releasing hormone, RANKL/RANK, B Cell Receptor Signaling Pathway, and Ras Signaling Pathway. It also involved in some inflammatory signaling pathways like IL-2, IL-3, IL-5, PI3K-Akt, and Toll-like Receptor Signaling Pathway, and Ebola Virus pathway on host (Fig. 7 and Supplementary file, Table S10).
Analysis of the ingredient-target interaction network and molecular docking
Based on the above analysis, we constructed an “ingredient-target interaction” network, showing relationships of “Chinese herbal-active ingredient-effector target-biological process-signaling pathway” (Fig. 8). In this network, 5 rectangles represent Chinese herbals, 18 triangles represent core potential active ingredients of XBJ, ellipses represent core top 10 potential effector targets of COVID-19, round rectangles represent core top 10 biological processes, hexagon represent core top 10 KEGG pathways, and octagons represent core top 10 Wikipathways. This indicated that the targets of the active ingredients of XBJ were distributed across different pathways and that XBJ might use multiple pathways to carry out its synergistic effects.
We applied molecular docking to assess protein-ligand binding potential, top 10 potential effector targets and their related 18 ingredients in XBJ and were selected. The molecular docking results showed that the 18 effective ingredients had a docking score of less than − 4.0 with those 10 targets, indicating they have potential binding activity. Among them, Ethyl ferulate-GAPDH, protocatechuic acid-ALB, Rutin-TNF, Apigenin-EGFR, Ethyl ferulate-MAPK1, Benzoylpaeoniflorin-CASP3, Cryptotanshinone-STAT3, Rosmarinic acid-MAPK8, Cryptotanshinone-PTGS2 and Salvianolic acid B-JUN had best binding activity than others (Table 3). Nine binding modes are displayed in Fig. 9. Therefore, these ingredients may be potential compounds in anti-COVID-19 in XBJ.
Discussion
XBJ has been approved for the treatment of severe infection (sepsis). For a long period of time in China, it was believed that XBJ could improve prognosis in severe lung infection [35] as well as 28-day mortality rate, Acute Physiologic Assessment and Chronic Health Evaluation (APACHE) II score, white blood cell (WBC) count, and temperature in sepsis patients without causing serious adverse events. A prospective, randomized controlled trial in 33 hospitals in China, published in September 2019, showed that XBJ can significantly improve the primary endpoint of pneumonia severity in patients with severe community-acquired pneumonia, as well as secondary endpoints such as mortality rate, mechanical-ventilation duration, and length of intensive-care unit (ICU) stay [36]. Since the start of the COVID-19 outbreak, XBJ has been recommended as a Chinese patent medicine in local COVID-19 diagnosis and treatment plans released by many provincial health commissions due to its rapid onset and significant efficacy in critically ill patients. This shows that XBJ might have important clinical value in COVID-19 treatment. However, the material bases and molecular effector mechanisms are still unclear. Therefore, analysis of the potential effector mechanisms of XBJ in the treatment of COVID-19, elucidating its potential active ingredients and their potential effector targets, and demonstrating the network effector mechanisms of XBJ on COVID-19, can provide a scientific basis for using XBJ in the clinical treatment of COVID-19.
This study preliminarily demonstrated XBJ’s “multiple component-multiple target-multiple pathway” effector characteristics, and our network topology analysis of potential effector targets identified some critical effector targets of the compound. GO and KEGG enrichment analyses found that the potential targets of XBJ involved multiple inflammation- and immune-related gene functions and signaling pathways, which might be one basis for XBJ treatment in COVID-19.
From a potential active-ingredient perspective, XBJ possesses potential anti-inflammatory and immune-boosting effects [37]. The three active ingredients of Carthamus tinctorius have protective effects in lipopolysaccharide (LPS)-induced acute lung injury (ALI) [38]. The potential anti-inflammatory components in XBJ inhibit NF-κB activity; decrease expression of TNF-α, IL-1β, and IL-6 [39]; alleviate inflammatory responses; and inhibit secretion of pro-inflammatory cytokines mediated by the high-mobility group box 1 protein (HMGB1)-receptor for advanced glycation endproducts (RAGE) axis, thereby decreasing mortality rate in a mouse model [40]. XBJ also upregulates toll-interacting proteins in septic rats to protect the lungs from permeable leakage and injury [41]. In addition, it prevents cytokine storm, inhibits inflammatory responses, and regulates regulatory T-cell (Treg)-T helper 17 cell (Th17) balance to improve survival in septic shock [42]. In addition, XBJ promotes the expression of annexin A1 to inhibit cleavage of pro-inflammatory cytokines and decreases IL-8 and TNF-α levels to protect rats from damage due to Acinetobacter baumannii sepsis [43]. SARS-CoV-2 replicates in respiratory-tract epithelial cells to cause acute inflammation and severe respiratory disease. During infection, local production of pro-inflammatory cytokines exacerbates disease progression. Therefore, the anti-inflammatory activity and cytokine-inhibitory effects of XBJ might constitute its potential mechanism in COVID-19 treatment.
In GO BP analysis, the 144 potential therapeutic targets of XBJ involved multiple inflammation- and immune-related BPs such as extracellular signal-regulated kinase 1 and 2 (ERK1, ERK2) cascade, the T-cell receptor signaling pathway, activation of MAPK activity, and cellular response to LPS. This suggests that the significant anti-inflammatory effects of XBJ are its therapeutic effects in inflammation. Virus-host interactions are an important aspect of viral replication. Ribonucleic acid (RNA) viruses such as influenza, Ebola virus, and SARS-CoV can induce Raf–mitogen-activated protein kinase kinase (MEK)–ERK signal transduction in the MAPK cascade, which is associated with replication of pathogenic RNA viruses in humans and allows for cell differentiation and proliferation [44]. This is consistent with the fact that XBJ targets the ERK1/ERK2 cascade.
Extensive proteinoid and serous exudates are present in the alveoli of COVID-19 patients. These cases also present bilateral diffuse alveolar damage accompanied by fibromyxoid exudates, and both lungs show apparent pulmonary edema, alveolar epithelial detachment, and hyaline-membrane formation [45]. In terms of infection-related serum markers, studies show that C-reactive protein (CRP), IL-6, and erythrocyte sedimentation rate (ESR) are significantly increased in many patients [46]. Severe cytokine storm can appear in severely to critically ill patients, resulting in excessive immune activation and excess production of IL-7, IL-10, granulocyte colony-stimulating factor (GCSF), interferon gamma inducible protein 10 kD (IP-10), monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1A (MIP1A), and TNF-α [47]. From this, it can be seen that SARS-CoV-2–mediated inflammation plays an important role in COVID-19 progression, and uncontrollable lung inflammation may be the main cause of death in COVID-19. Therefore, intervention measures to reduce inflammation might help decrease the mortality rate [48].
With regard to effector targets, the degree values of GAPDH, albumin (ALB), TNF, EGFR, MAPK1, CASP3, STAT3, MAPK8, PTGS2, JUN, IL-2, Estrogen Receptor 1 (ESR1), and MAPK14 were all > 40, suggesting that in COVID-19 these could be the main therapeutic targets of XBJ’s active ingredients. Then we verified the binding activity of the XBJ key compounds and the COVID-19 key targets via molecular docking. The results indicated that they have potential binding activity and these ingredients may be potential compounds in anti-COVID-19 of XBJ. Wikipathways and KEGG enrichment analysis contained 40 and 94 signaling pathways, showing that the 144 potential XBJ therapeutic targets were involved in many inflammation- and immune-related signaling pathways.
The renin-angiotensin system (RAS), OS and cell death, cytokine storm, and endothelial dysfunction are four main pathways in COVID-19 pathogenesis. ACE is a receptor in the airway, alveoli, and vascular endothelium. COVID-19 uses ACE to enter type II pneumocytes or intestinal epithelial cells in order to induce ACE2 internalization and shedding, resulting in the occurrence and development of acute respiratory distress syndrome (ARDS) [49]. Many of XBJ’s active ingredients act on multiple targets in the RAS, which could potentially interfere with ACE receptors. NF-κB activation exacerbates lung inflammation caused by SARS-CoV infection, and inhibition of NF-κB signaling significantly reduces such inflammation and increases the survival rate of SARS-CoV-infected mice [50, 51]. In addition, NF-κB is an important transcription factor that induces expression of viral genes, and inhibition of NF-κB activation is an immune evasion mechanism of SARS-CoV [52]. Therefore, we speculate that XBJ’s active ingredients might interfere with the NF-κB pathway and regulate innate immunity and inflammation during viral infection to alleviate lung inflammation during COVID-19.
One study has shown that XBJ inhibits MAPK and NF-κB expression and has protective effects in ALI [53]. The compound regulates the NF-κB, MAPK, and PI3K–Akt pathways in mouse macrophages and downregulates inflammatory cytokines such as IL-6, TNF-α, MCP-1, MIP-2, and serum IL-10 to increase the survival rate of septic mice [15]. XBJ downregulates toll-like receptor 4 (TLR-4) and NF-κB expression to carry out its anti-inflammatory effects. MAPK activation can promote the expression and release of pro-inflammatory cytokines such as TNF-α and IL-1β, − 6, and − 8; it is a core factor in inflammation regulation. Viruses usually directly or indirectly affect the PI3K–Akt pathway to control intracellular-signaling pathways. EGFR aggregation and binding of influenza virus to cell surfaces might activate Akt. The PI3K–Akt signaling pathway might synergize with the RAS to promote viral entry, which has significant effects in viral infection in humans [54]. TLR-2, TLR-3, and TLR-4 activation by COVID-19 causes the release of inflammatory cytokines such as IL-1β. The binding of SARS-CoV-2 to TLRs causes release of pro-IL-1β, inflammasome activation, and production of mature IL-1β. Pro-inflammatory cytokines are important mediators of local and systemic inflammation. Viral particles first invade the respiratory mucosa before infecting other cells, thereby inducing a series of immune responses leading to cytokine storm [55]. Therefore, XBJ could be used to treat COVID-19 patients due to its anti-inflammatory effects, anti-immune apoptosis, and alleviation of pneumonia-induced multi-organ damage. In addition, we found that critical nodes on our “XBJ active-compound-potential effector target” network analysis map participated in the aforementioned pathways, suggesting that the predictions made in this study are somewhat accurate.
When compared to other research works on NP related to COVID-19 prevention and treatment, some of our research results and conclusions are consistent with the current similar research in some aspects. Qingfei Paidu Decoction (QFPD), a clinically used Chinese medicine for treating COVID-19 patients in China, has been shown by a recent NP research that the therapeutic effects of QFPD against COVID-19 may be attributed to the anti-inflammatory effects related to the thrombin and TLR signaling pathway [56]. What’s more, QFPD could protect COVID-19 injury via anti-Viral, anti-Inflammatory activity and metabolic programming [57]. Another research shows that Pudilan (PDL), clinically used as an anti-SARS-CoV-2 infective agent in China. PDL might moderate the immune system to shorten the course of the disease, delay disease progression, and reduce the mortality rate [58]. Hence, QFPD and PDL, together with XBJ may have a therapeutic effect on COVID-19 through three aspects, including the immune system, anti-inflammation, and anti-virus entry into cells.
Previous similar study have shown that 8 key compounds and 15 key targets of XBJ were screened and verified by molecular docking [59], on the basis of this research, we have made improvements in the criteria for screening active ingredients, drug targets, and disease targets (Table 4). As XBJ is administered intravenously, which does not need to be absorbed through the gastrointestinal tract, therefore, we selected active ingredients of XBJ that had been detected by LC-MS in our study. What’s more, we used the latest update version of Swiss database to predict the targets of XBJ. In terms of the potential core targets of COVID-19, we finally found different results from previous studies.
Compared with previous similar study, there is a significant advantage of our study. To further verify our analysis results, we found that the 451 targets of XBJ (Supplementary file, Table S1) and 332 putative COVID-19-interacting human protein had fourteen intersections: BRD2, BRD4, IMPDH2, POLA1, ATP6AP1, SIGMAR1, HMOX1, DNMT1, HDAC2, PABPC1, EIF4H, PRKACA, ABCC1, and COMT.
It should be noted that BRD2/BRD4 are potential drug targets of Envelope (E) protein, a structural proteins of COVID-19, which plays a central role in virus morphogenesis and assembly. BRD2/BRD4 (Bromodomain extra terminal proteins) are implicated as epigenetic factors that regulate genes crucial for cell cycle progression, inflammation and immune response. What’s more, E binds to protein M, and co-expression of M and E is sufficient for VLP formation and release. Lack of E reduces viral titers about 20-fold [14]. From this speculation, XBJ might help the treatment of COVID-19 by disrupting the interaction between protein E and BRDs (Fig. 10).
However, there are several limitations in the current study. The host proteins and existing targets of the relevant coronaviruses are used to be potential therapeutic targets for COVID-19. Although SARS-CoV-2 shared high nucleotide sequence identity with other HCoVs, our predictions are not SARS-CoV-2 specific by lack of the known host proteins on SARS-CoV-2. In terms of COVID-19 treatment targets, we have an adopted alternative strategy that is currently achievable.
SARS is associated with epithelial-cell proliferation and an increase in macrophages in the lung [60]. And diffuse alveolar damage was seen in COVID-19 cases [61]. Regarding the differences between COVID-19 and SARS. Studies reveal that SARS-CoV-2 is very similar in structure and pathogenicity with SARS-CoV, but the most important structural protein, i.e., the spike protein (S), is slightly different in these viruses. Compared to other beta coronaviruses, the presence of a furin-like cleavage site in SARS-CoV-2 facilitates the S protein priming and might increase the efficiency of the spread of SARS-CoV-2 [62, 63]. COVID-19 has diverse epidemiological and biological characteristics, making it more contagious than SARS-CoV and MERS-CoV. It has affected more people in a short time period compared to SARS-CoV and MERS-CoV, although the fatality rate of MERS-CoV was higher than SARS-CoV and SARS-CoV [64].
We explore the molecular mechanism of Xuebijing injection in COVID-19 based on the premise of host and protein interaction. We use coronavirus-related host proteins as potential targets, which aim to produce an indirect intervention against viral targets for the treatment of COVID-19.
Conclusions
XBJ could regulate different genes, act on different pathways, and synergize anti-inflammatory and immunoregulatory effects in COVID-19. This demonstrates the synergy of multiple targets and pathways among different components, as well as the holistic concept of TCM formulations.
Availability of data and materials
All data are available in the manuscript and are shown in tables, figures, and supplemental files.
References
Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395(10223):470–3. https://doi.org/10.1016/S0140-6736(20)30185-9.
Lu H. Drug treatment options for the 2019-new coronavirus (2019-nCoV). Biosci Trends. 2020;14(1):69–71. https://doi.org/10.5582/bst.2020.01020.
Li H, Zhou Y, Zhang M, Wang H, Zhao Q, Liu J. Updated Approaches against SARS-CoV-2. Antimicrob Agents Chemother. 2020;64(6). https://doi.org/10.1128/AAC.00483-20.
Ren JL, Zhang AH, Wang XJ. Traditional Chinese medicine for COVID-19 treatment. Pharmacol Res. 2020;155:104743. https://doi.org/10.1016/j.phrs.2020.104743.
Zhang Q, Li J, Liang X, Xie H, Sun H, Lin X, et al. The preventive effect of Chinese herbal preparation Xuebijing against hyperactive inflammation after hepato-pancreato-biliary surgery. Ann Transl Med. 2019;7(18):481. https://doi.org/10.21037/atm.2019.07.78.
He F, Wang J, Liu Y, Wang X, Cai N, Wu C, et al. Xuebijing injection induces anti-inflammatory-like effects and downregulates the expression of TLR4 and NF-kappaB in lung injury caused by dichlorvos poisoning. Biomed Pharmacother. 2018;106:1404–11. https://doi.org/10.1016/j.biopha.2018.07.111.
Yin Q, Li C. Treatment effects of xuebijing injection in severe septic patients with disseminated intravascular coagulation. Evid Based Complement Alternat Med. 2014;2014:949254. https://doi.org/10.1155/2014/949254.
Cheng C, Lin JZ, Li L, Yang JL, Jia WW, Huang YH, et al. Pharmacokinetics and disposition of monoterpene glycosides derived from Paeonia lactiflora roots (Chishao) after intravenous dosing of antiseptic XueBiJing injection in human subjects and rats. Acta Pharmacol Sin. 2016;37(4):530–44. https://doi.org/10.1038/aps.2015.103.
Liu MW, Liu R, Wu HY, Zhang W, Xia J, Dong MN, et al. Protective effect of Xuebijing injection on D-galactosamine- and lipopolysaccharide-induced acute liver injury in rats through the regulation of p38 MAPK, MMP-9 and HO-1 expression by increasing TIPE2 expression. Int J Mol Med. 2016;38(5):1419–32. https://doi.org/10.3892/ijmm.2016.2749.
Chen Y, Tong H, Pan Z, Jiang D, Zhang X, Qiu J, et al. Xuebijing injection attenuates pulmonary injury by reducing oxidative stress and proinflammatory damage in rats with heat stroke. Exp Ther Med. 2017;13(6):3408–16. https://doi.org/10.3892/etm.2017.4444.
Li T, Qian Y, Miao Z, Zheng P, Shi T, Jiang X, et al. Xuebijing injection alleviates Pam3CSK4-induced inflammatory response and protects mice from Sepsis caused by methicillin-resistant Staphylococcus aureus. Front Pharmacol. 2020;11:104. https://doi.org/10.3389/fphar.2020.00104.
Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–90. https://doi.org/10.1038/nchembio.118.
Li S, Zhang B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin J Nat Med. 2013;11(2):110–20. https://doi.org/10.1016/S1875-5364(13)60037-0.
Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020. https://doi.org/10.1038/s41586-020-2332-7.
Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. https://doi.org/10.1038/s41421-020-0153-3.
Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–3. https://doi.org/10.1038/s41586-020-2012-7.
Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–9. https://doi.org/10.1038/s41586-020-2008-3.
Zumla A, Chan JF, Azhar EI, Hui DS, Yuen KY. Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016;15(5):327–47. https://doi.org/10.1038/nrd.2015.37.
Messina F, Giombini E, Agrati C, Vairo F, Ascoli Bartoli T, Al Moghazi S, et al. COVID-19: viral-host interactome analyzed by network based-approach model to study pathogenesis of SARS-CoV-2 infection. J Transl Med. 2020;18(1):233. https://doi.org/10.1186/s12967-020-02405-w.
Daina A, Michielin O, Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W64. https://doi.org/10.1093/nar/gkz382.
Rappaport N, Fishilevich S, Nudel R, Twik M, Belinky F, Plaschkes I, et al. Rational confederation of genes and diseases: NGS interpretation via GeneCards, MalaCards and VarElect. Biomed Eng Online. 2017;16(Suppl 1):72. https://doi.org/10.1186/s12938-017-0359-2.
Hoffmann M, Kleine-Weber H, Schroeder S, Kruger N, Herrler T, Erichsen S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–80 e8. https://doi.org/10.1016/j.cell.2020.02.052.
Wang J, Zhao S, Liu M, Zhao Z, Xu Y, Wang P, et al. ACE2 expression by colonic epithelial cells is associated with viral infection, immunity and energy metabolism. medRxiv. 2020:2020.02.05.20020545. https://doi.org/10.1101/2020.02.05.20020545.
Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020. https://doi.org/10.1038/s41586-020-2286-9.
Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics. 2014;47(8 13):1–24. https://doi.org/10.1002/0471250953.bi0813s47.
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D8. https://doi.org/10.1093/nar/gkw937.
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8 Suppl 4:S11. https://doi.org/10.1186/1752-0509-8-S4-S11.
Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7. https://doi.org/10.1089/omi.2011.0118.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. https://doi.org/10.1038/75556.
Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–80. https://doi.org/10.1093/nar/gkh063.
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25(8):1091–3. https://doi.org/10.1093/bioinformatics/btp101.
Bindea G, Galon J, Mlecnik B. CluePedia Cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29(5):661–3. https://doi.org/10.1093/bioinformatics/btt019.
Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–61. https://doi.org/10.1002/jcc.21334.
Zuo L, Sun Z, Hu Y, Sun Y, Xue W, Zhou L, et al. Rapid determination of 30 bioactive constituents in XueBiJing injection using ultra high performance liquid chromatography-high resolution hybrid quadrupole-orbitrap mass spectrometry coupled with principal component analysis. J Pharm Biomed Anal. 2017;137:220–8. https://doi.org/10.1016/j.jpba.2017.01.024.
Li C, Wang P, Zhang L, Li M, Lei X, Liu S, et al. Efficacy and safety of Xuebijing injection (a Chinese patent) for sepsis: a meta-analysis of randomized controlled trials. J Ethnopharmacol. 2018;224:512–21. https://doi.org/10.1016/j.jep.2018.05.043.
Song Y, Yao C, Yao Y, Han H, Zhao X, Yu K, et al. XueBiJing injection versus placebo for critically ill patients with severe community-acquired pneumonia: a randomized controlled trial. Crit Care Med. 2019;47(9):e735–e43. https://doi.org/10.1097/CCM.0000000000003842.
Liu Y, Tian X, Cui M, Zhao S. Safflower yellow inhibits angiotensin II-induced adventitial fibroblast proliferation and migration. J Pharmacol Sci. 2014;126(2):107–14. https://doi.org/10.1254/jphs.14055fp.
Wang YP, Guo Y, Wen PS, Zhao ZZ, Xie J, Yang K, et al. Three ingredients of safflower alleviate acute lung injury and inhibit NET release induced by lipopolysaccharide. Mediat Inflamm. 2020;2020:2720369. https://doi.org/10.1155/2020/2720369.
Jiang M, Zhou M, Han Y, Xing L, Zhao H, Dong L, et al. Identification of NF-kappaB inhibitors in Xuebijing injection for sepsis treatment based on bioactivity-integrated UPLC-Q/TOF. J Ethnopharmacol. 2013;147(2):426–33. https://doi.org/10.1016/j.jep.2013.03.032.
Wang Q, Wu X, Tong X, Zhang Z, Xu B, Zhou W. Xuebijing ameliorates Sepsis-induced lung injury by Downregulating HMGB1 and RAGE expressions in mice. Evid Based Complement Alternat Med. 2015;2015:860259. https://doi.org/10.1155/2015/860259.
Liu MW, Wang YH, Qian CY, Li H. Xuebijing exerts protective effects on lung permeability leakage and lung injury by upregulating toll-interacting protein expression in rats with sepsis. Int J Mol Med. 2014;34(6):1492–504. https://doi.org/10.3892/ijmm.2014.1943.
Chen X, Feng Y, Shen X, Pan G, Fan G, Gao X, et al. Anti-sepsis protection of Xuebijing injection is mediated by differential regulation of pro- and anti-inflammatory Th17 and T regulatory cells in a murine model of polymicrobial sepsis. J Ethnopharmacol. 2018;211:358–65. https://doi.org/10.1016/j.jep.2017.10.001.
He XD, Wang Y, Wu Q, Wang HX, Chen ZD, Zheng RS, et al. Xuebijing protects rats from Sepsis challenged with Acinetobacter baumannii by promoting Annexin A1 expression and inhibiting Proinflammatory cytokines secretion. Evid Based Complement Alternat Med. 2013;2013:804940. https://doi.org/10.1155/2013/804940.
Pleschka S. RNA viruses and the mitogenic Raf/MEK/ERK signal transduction cascade. Biol Chem. 2008;389(10):1273–82. https://doi.org/10.1515/BC.2008.145.
Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8(4):420–2. https://doi.org/10.1016/S2213-2600(20)30076-X.
Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507–13. https://doi.org/10.1016/S0140-6736(20)30211-7.
Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. https://doi.org/10.1016/S0140-6736(20)30183-5.
Fu Y, Cheng Y, Wu Y. Understanding SARS-CoV-2-mediated inflammatory responses: from mechanisms to potential therapeutic tools. Virol Sin. 2020. https://doi.org/10.1007/s12250-020-00207-4.
Sparks MA, South A, Welling P, Luther JM, Cohen J, Byrd JB, et al. Sound science before quick Judgement regarding RAS blockade in COVID-19. Clin J Am Soc Nephrol. 2020;15(5):714–6. https://doi.org/10.2215/CJN.03530320.
Canton J, Fehr AR, Fernandez-Delgado R, Gutierrez-Alvarez FJ, Sanchez-Aparicio MT, Garcia-Sastre A, et al. MERS-CoV 4b protein interferes with the NF-kappaB-dependent innate immune response during infection. PLoS Pathog. 2018;14(1):e1006838. https://doi.org/10.1371/journal.ppat.1006838.
DeDiego ML, Nieto-Torres JL, Regla-Nava JA, Jimenez-Guardeno JM, Fernandez-Delgado R, Fett C, et al. Inhibition of NF-kappaB-mediated inflammation in severe acute respiratory syndrome coronavirus-infected mice increases survival. J Virol. 2014;88(2):913–24. https://doi.org/10.1128/JVI.02576-13.
Deng L, Zeng Q, Wang M, Cheng A, Jia R, Chen S, et al. Suppression of NF-kappaB Activity: A Viral Immune Evasion Mechanism. Viruses. 2018;10(8). https://doi.org/10.3390/v10080409.
Liu MW, Su MX, Zhang W, Wang YQ, Chen M, Wang L, et al. Protective effect of Xuebijing injection on paraquat-induced pulmonary injury via down-regulating the expression of p38 MAPK in rats. BMC Complement Altern Med. 2014;14:498. https://doi.org/10.1186/1472-6882-14-498.
Dunn EF, Connor JH. HijAkt: the PI3K/Akt pathway in virus replication and pathogenesis. Prog Mol Biol Transl Sci. 2012;106:223–50. https://doi.org/10.1016/B978-0-12-396456-4.00002-X.
Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak - an update on the status. Mil Med Res. 2020;7(1):11. https://doi.org/10.1186/s40779-020-00240-0.
Yang R, Liu H, Bai C, Wang Y, Zhang X, Guo R, et al. Chemical composition and pharmacological mechanism of Qingfei Paidu decoction and ma Xing Shi Gan decoction against coronavirus disease 2019 (COVID-19): in silico and experimental study. Pharmacol Res. 2020;157:104820. https://doi.org/10.1016/j.phrs.2020.104820.
Chen J, Wang YK, Gao Y, Hu LS, Yang JW, Wang JR, et al. Protection against COVID-19 injury by qingfei paidu decoction via anti-viral, anti-inflammatory activity and metabolic programming. Biomed Pharmacother. 2020;129:110281. https://doi.org/10.1016/j.biopha.2020.110281.
Kong Q, Wu Y, Gu Y, Lv Q, Qi F, Gong S, et al. Analysis of the molecular mechanism of Pudilan (PDL) treatment for COVID-19 by network pharmacology tools. Biomed Pharmacother. 2020;128:110316. https://doi.org/10.1016/j.biopha.2020.110316.
Zhang Y-L, Cui Q, Zhang D, Ma X, Zhang G-W. Efficacy of Xuebijing injection for the treatment of coronavirus disease 2019 via network pharmacology. Traditional Medicine Research. 2020;5(4):201–15. https://doi.org/10.12032/tmr20200507178.
Nicholls JM, Poon LL, Lee KC, Ng WF, Lai ST, Leung CY, et al. Lung pathology of fatal severe acute respiratory syndrome. Lancet. 2003;361(9371):1773–8. https://doi.org/10.1016/s0140-6736(03)13413-7.
Sauter JL, Baine MK, Butnor KJ, Buonocore DJ, Chang JC, Jungbluth AA, et al. Insights into pathogenesis of fatal COVID-19 pneumonia from histopathology with immunohistochemical and viral RNA studies. Histopathology. 2020. https://doi.org/10.1111/his.14201.
Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antivir Res. 2020;176:104742. https://doi.org/10.1016/j.antiviral.2020.104742.
Millet JK, Whittaker GR. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. Proc Natl Acad Sci U S A. 2014;111(42):15214–9. https://doi.org/10.1073/pnas.1407087111.
Meo SA, Alhowikan AM, Al-Khlaiwi T, Meo IM, Halepoto DM, Iqbal M, et al. Novel coronavirus 2019-nCoV: prevalence, biological and clinical characteristics comparison with SARS-CoV and MERS-CoV. Eur Rev Med Pharmacol Sci. 2020;24(4):2012–9. https://doi.org/10.26355/eurrev_202002_20379.
Acknowledgements
We thank LetPub for its linguistic assistance during the preparation of this manuscript.
Funding
This research was funded by grants from the National Natural Science Foundation of China (No.81973814), the Liu Xiaohong Famous Traditional Chinese Medicine Inheritance Studio from the Traditional Chinese Medicine Bureau of Guangdong Province (No.201805), the Construction Project of the National Clinical Medical Research Center (respiratory department, No.2110200309), the Construction Project of High Level Hospital on Modified Weijing Granules (No.211010010603), and the Student Learning Team Incubation Project of Innovation Academy from The First Affiliated Hospital of Guangzhou University of Chinese Medicine (No.2018XXTD003), and the Guangdong Provincial Science and Technology Plan Project (South China Traditional Chinese Medicine Collaborative Innovation Center, No.2014b0902002), 2018 Guangzhou University of Chinese Medicine National University Student Innovation and Entrepreneurship Training Project (No.201810572038), and 2020 National College Student Innovation and Entrepreneurship Training Project of Guangzhou University of Chinese Medicine (No. 202010572001).
Author information
Authors and Affiliations
Contributions
Conceptualization, LXH, and JY; methodology, ZWJ, and YQ; software, NYS; formal analysis, ZSF and YLL; investigation, ZWJ; resources, ZHF; data curation, NYS; writing-original draft preparation, ZWJ, and YQ; writing-review and editing, JY; supervision, LXH; project administration, JY; funding acquisition, LXH. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable. This manuscript does not report on or involve the use of any animal or human data or tissue.
Consent for publication
Not applicable. This manuscript does not contain data from any individual person.
Competing interests
The authors declare that they have no conflicts of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
The detailed targets information. Table S2. 451drug targets of XBJ. Table S3. 251 coronavirus pneumonia targets. Table S4. 119 HCoV-associated host proteins. Table S5. 3525 genes co-expressed with ACE2. Table S6. 144 potential COVID-19 targets. Table S7. Network topology analysis results. Table S4. 119 HCoV-associated host proteins. Table S8. GO-BP results. Table S9. KEGG-pathways results. Table S10. Wiki-pathways results.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Zheng, Wj., Yan, Q., Ni, Ys. et al. Examining the effector mechanisms of Xuebijing injection on COVID-19 based on network pharmacology. BioData Mining 13, 17 (2020). https://doi.org/10.1186/s13040-020-00227-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13040-020-00227-6