Abstract
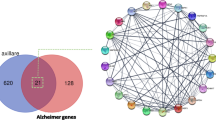
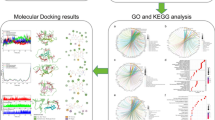
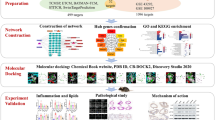
Scientific research has demonstrated that Tinospora cordifolia acts as an anti-aging agent in several experimental models, generating global interest in its underlying molecular mechanisms of this activity. The aim of the study was to identify the possible phytochemical compounds of T. cordifolia that might combat age-related illness through integrating network pharmacology, molecular docking techniques, and molecular dynamics (MD) study to explore their potential mechanisms of action. To carry out this study, several databases were used, including PubChem, KNApSAcK family database, PubMed, SwissADME, Molsoft, SwissTargetPrediction, GeneCards, and OMIM database. For network development and GO enrichment analysis KEGG, ShinyGo 0.77, and the STRING database were used. For better analysis, the networks were also constructed using Cytoscape 3.9.1. The Cytoscape network analyzer tool was used for data analysis, and molecular docking was done via Vina-GPU-2.0. The best compounds and AKT1 were finally subjected to MD simulation for 100 ns. The CytoHubba plugin of Cytoscape identified ten key targets, commonly called hub genes, including AKT1, GAPDH, and TP53, and so on. GO and KEGG pathway enrichment analysis revealed the relevant biological processes, cellular components, and molecular functions involved in treating aging-related disorders. KEGG pathway analysis involved neuroactive ligand–receptor interactions, lipid and atherosclerosis, and cAMP signaling. The docking of 100 T. cordifolia compounds with AKT1 demonstrated good binding affinity, particularly for Amritoside, Sitagliptin, Berberine, and Piperine. Finally, the relative stability of four-hit phytochemicals was validated by MD simulation, which may be the most crucial compound for anti-aging activity. In conclusion, this study used network pharmacology, molecular docking, and MD simulation to identify the compounds in T. cordifolia and proposed a potential mechanism for anti-aging activity. These results suggest future directions for the prevention and treatment of age-related diseases.












Similar content being viewed by others
References
Dudhamal T (2014) Shalya Tantra—the Ayurvedic surgical branch—a literary review. OIIRJ 4:197–207
Corrêa RCG, Peralta RM, Haminiuk CWI et al (2018) New phytochemicals as potential human anti-aging compounds: reality, promise, and challenges. Crit Rev Food Sci Nutr 58:942–957. https://doi.org/10.1080/10408398.2016.1233860
Lenart P, Krejci L (2016) DNA, the central molecule of aging. Mutat Res Mol Mech Mutagen 786:1–7. https://doi.org/10.1016/j.mrfmmm.2016.01.007
López-Otín C, Blasco MA, Partridge L et al (2013) The hallmarks of aging. Cell 153:1194–1217. https://doi.org/10.1016/j.cell.2013.05.039
de Almeida AJPO, Ribeiro TP, de Medeiros IA (2017) Aging: molecular pathways and implications on the cardiovascular system. Oxid Med Cell Longev 2017:e7941563. https://doi.org/10.1155/2017/7941563
Ding A-J, Zheng S-Q, Huang X-B et al (2017) Current perspective in the discovery of anti-aging agents from natural products. Nat Prod Bioprospect 7:335–404. https://doi.org/10.1007/s13659-017-0135-9
Fuentealba M, Dönertaş HM, Williams R et al (2019) Using the drug–protein interactome to identify anti-ageing compounds for humans. PLoS Comput Biol 15:e1006639. https://doi.org/10.1371/journal.pcbi.1006639
Liu Y, Liu Y, Zhang W et al (2020) Network pharmacology-based strategy to investigate the pharmacological mechanisms of Ginkgo biloba extract for aging. Evid Based Complement Altern Med 2020:8508491. https://doi.org/10.1155/2020/8508491
Adhikari SR, Pokhrel K (2019) The medicinal uses of Tinospora cordifolia (Gurjo). Himal Biodivers. https://doi.org/10.3126/hebids.v7i1.40190
Banerjee A, Pavane MS, Banu LH et al (2021) Traditional medicine for aging-related disorders: implications for drug discovery. In: Stem cells and aging. Elsevier, Amsterdam, pp 281–297
Saxena C, Rawat G (2019) Tinospora cordifolia (Giloy)—therapeutic uses and importance: a review. Curr Res Pharm Sci 9:42–45. https://doi.org/10.24092/CRPS.2019.090302
Ghosh S, Saha S (2012) Tinospora cordifolia: one plant, many roles. Anc Sci Life 31:151. https://doi.org/10.4103/0257-7941.107344
Hegde S, Jayaraj M (2016) A review of the medicinal properties, phytochemical and biological active compounds of Tinospora sinensis (Lour.) Merr. J Biol Act Prod Nat 6:84–94. https://doi.org/10.1080/22311866.2016.1185968
Khare CP (2007) Indian medicinal plants: an illustrated dictionary. https://doi.org/10.1007/978-0-387-70638-2
Pathak P, Vyas M, Vyas H, Naria M (2016) Rasayana effect of Guduchi Churna on the life span of Drosophila melanogaster. Ayu 37:67–70. https://doi.org/10.4103/ayu.AYU_11_16
Bhandari A, Kalotra S, Bajaj P et al (2022) Dietary intervention with Tinospora cordifolia improved aging-related decline in locomotor coordination and cerebellar cell survival and plasticity in female rats. Biogerontology 23:809–824. https://doi.org/10.1007/s10522-022-09975-w
Bhandari A, Sharma AK, Singh H et al (2022) Aging-related changes in metabolic indicators in female rats and their management with Tinospora cordifolia. Biogerontology 23:363–380. https://doi.org/10.1007/s10522-022-09962-1
Khanal P, Patil BM, Mandar BK et al (2019) Network pharmacology-based assessment to elucidate the molecular mechanism of anti-diabetic action of Tinospora cordifolia. Clin Phytosci 5:35. https://doi.org/10.1186/s40816-019-0131-1
Liu W, Fan Y, Tian C et al (2020) Deciphering the molecular targets and mechanisms of HGWD in the treatment of rheumatoid arthritis via network pharmacology and molecular docking. Evid Based Complement Altern Med 2020:e7151634. https://doi.org/10.1155/2020/7151634
Guo P, Cai C, Wu X et al (2019) An insight into the molecular mechanism of berberine towards multiple cancer types through systems pharmacology. Front Pharmacol 10:857. https://doi.org/10.3389/fphar.2019.00857
Wu Q, Chen Y, Gu Y et al (2020) Systems pharmacology-based approach to investigate the mechanisms of Danggui-Shaoyao-san prescription for treatment of Alzheimer’s disease. BMC Complement Med Ther 20:282. https://doi.org/10.1186/s12906-020-03066-4
Hopkins AL (2008) Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol 4:682–690. https://doi.org/10.1038/nchembio.118
Lan S, Duan J, Zeng N et al (2021) Network pharmacology-based screening of the active ingredients and mechanisms of Huangqi against aging. Medicine (Baltim) 100:e25660. https://doi.org/10.1097/MD.0000000000025660
Li S, Zhang B (2013) Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin J Nat Med 11:110–120. https://doi.org/10.1016/S1875-5364(13)60037-0
Luo T-T, Lu Y, Yan S-K et al (2020) Network pharmacology in research of Chinese medicine formula: methodology, application and prospective. Chin J Integr Med 26:72–80. https://doi.org/10.1007/s11655-019-3064-0
Chandran U, Mehendale N, Patil S et al (2017) Network pharmacology. In: Innovative approaches in drug discovery. Elsevier, Amsterdam, pp 127–164
Gong P, Wang D, Cui D et al (2021) Anti-aging function and molecular mechanism of Radix Astragali and Radix Astragali preparata via network pharmacology and PI3K/Akt signaling pathway. Phytomedicine 84:153509. https://doi.org/10.1016/j.phymed.2021.153509
Afendi FM, Okada T, Yamazaki M et al (2012) KNApSAcK family databases: integrated metabolite–plant species databases for multifaceted plant research. Plant Cell Physiol 53:e1. https://doi.org/10.1093/pcp/pcr165
Bala M, Verma PK, Awasthi S et al (2015) Chemical prospection of important Ayurvedic plant Tinospora cordifolia by UPLC–DAD–ESI–QTOF-MS/MS and NMR. Nat Prod Commun 10:43–48
Chi S, She G, Han D et al (2016) Genus Tinospora: ethnopharmacology, phytochemistry, and pharmacology. Evid Based Complement Altern Med 2016:e9232593. https://doi.org/10.1155/2016/9232593
Mandar BK, Khanal P, Patil BM et al (2021) In silico analysis of phytoconstituents from Tinospora cordifolia with targets related to diabetes and obesity. In Silico Pharmacol 9:3. https://doi.org/10.1007/s40203-020-00063-w
Modi B, Koirala N, Aryal SP et al (2021) Tinospora cordifolia (Willd.) Miers: phytochemical composition, cytotoxicity, proximate analysis and their biological activities. Cell Mol Biol 67:50–57. https://doi.org/10.14715/cmb/2021.67.1.8
Sharma P, Dwivedee BP, Bisht D et al (2019) The chemical constituents and diverse pharmacological importance of Tinospora cordifolia. Heliyon 5:e02437. https://doi.org/10.1016/j.heliyon.2019.e02437
Singh D, Chaudhuri PK (2017) Chemistry and pharmacology of Tinospora cordifolia. Nat Prod Commun 12:299–308
Upadhyay A, Kumar K, Kumar A, Mishra H (2010) Tinospora cordifolia (Willd.) Hook. f. and Thoms. (Guduchi)—validation of the Ayurvedic pharmacology through experimental and clinical studies. Int J Ayurveda Res 1:112. https://doi.org/10.4103/0974-7788.64405
Daina A, Michielin O, Zoete V (2017) SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep 7:42717. https://doi.org/10.1038/srep42717
Daina A, Michielin O, Zoete V (2019) SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 47:W357–W364. https://doi.org/10.1093/nar/gkz382
Stelzer G, Rosen N, Plaschkes I et al (2016) The GeneCards Suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinform. https://doi.org/10.1002/cpbi.5
Amberger J, Bocchini CA, Scott AF, Hamosh A (2009) McKusick’s Online Mendelian Inheritance in Man (OMIM(R)). Nucleic Acids Res 37:D793–D796. https://doi.org/10.1093/nar/gkn665
Hamosh A (2004) Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res 33:D514–D517. https://doi.org/10.1093/nar/gki033
Szklarczyk D, Gable AL, Lyon D et al (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47:D607–D613. https://doi.org/10.1093/nar/gky1131
Shannon P, Markiel A, Ozier O et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Ge Q, Chen L, Yuan Y et al (2020) Network pharmacology-based dissection of the anti-diabetic mechanism of Lobelia chinensis. Front Pharmacol 11:347. https://doi.org/10.3389/fphar.2020.00347
Burley SK, Bhikadiya C, Bi C et al (2021) RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res 49:D437–D451. https://doi.org/10.1093/nar/gkaa1038
Valdés-Tresanco M, Valdés-Tresanco M, Valiente P, Moreno Frias E (2020) AMDock: a versatile graphical tool for assisting molecular docking with AutoDock Vina and AutoDock4. Biol Direct 15:12. https://doi.org/10.1186/s13062-020-00267-2
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 31:455–461. https://doi.org/10.1002/jcc.21334
Tian W, Chen C, Lei X et al (2018) CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res 46:W363–W367. https://doi.org/10.1093/nar/gky473
Morris GM, Lim-Wilby M (2008) Molecular docking. Methods Mol Biol (Clifton NJ) 443:365–382. https://doi.org/10.1007/978-1-59745-177-2_19
Warren GL, Andrews CW, Capelli A-M et al (2006) A critical assessment of docking programs and scoring functions. J Med Chem 49:5912–5931. https://doi.org/10.1021/jm050362n
Khanal P, Patil VS, Bhandare VV et al (2022) Computational investigation of benzalacetophenone derivatives against SARS-CoV-2 as potential multi-target bioactive compounds. Comput Biol Med 146:105668. https://doi.org/10.1016/j.compbiomed.2022.105668
Ding J, Tang S, Wang L et al (2022) Vina-GPU 2.0: further accelerating AutoDock Vina and its derivatives with GPUs. J Chem Inf Model 63(7):1982–1998
Pronk S, Páll S, Schulz R et al (2013) GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics (Oxf Engl) 29:845–854. https://doi.org/10.1093/bioinformatics/btt055
Vanommeslaeghe K, Hatcher E, Acharya C et al (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31:671–690. https://doi.org/10.1002/jcc.21367
Mi H, Muruganujan A, Casagrande JT, Thomas PD (2013) Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8:1551–1566. https://doi.org/10.1038/nprot.2013.092
Meng X-Y, Zhang H-X, Mezei M, Cui M (2011) Molecular Docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des 7:146–157
Nadkarni KM (1908) Indian Materia Medica, vol i
Mittal J (2014) Tinospora cordifolia: a multipurpose medicinal plant—a review. J Med Plants Stud 2:32–47
Tiwari P, Nayak P, Prusty SK, Sahu PK (2018) Phytochemistry and pharmacology of Tinospora cordifolia: a review. Syst Rev Pharm 9:70–78. https://doi.org/10.5530/srp.2018.1.14
Khanal P, Mandar BK et al (2019) In silico antidiabetic screening of borapetoside C, cordifolioside A and magnoflorine. Indian J Pharm Sci 81:550–555. https://doi.org/10.36468/pharmaceutical-sciences.543
Sharma R, Bolleddu R, Maji JK et al (2021) In vitro α-amylase, α-glucosidase inhibitory activities and in vivo anti-hyperglycemic potential of different dosage forms of Guduchi (Tinospora cordifolia [Willd.] Miers) prepared with Ayurvedic Bhavana Process. Front Pharmacol 12:642300
Patel MB, Mishra S (2011) Hypoglycemic activity of alkaloidal fraction of Tinospora cordifolia. Phytomed Int J Phytother Phytopharm 18:1045–1052. https://doi.org/10.1016/j.phymed.2011.05.006
de Oliveira AM, Conserva LM, De Souza Ferro JN et al (2012) Antinociceptive and anti-inflammatory effects of octacosanol from the leaves of Sabicea grisea var. grisea in mice. Int J Mol Sci 13:1598–1611. https://doi.org/10.3390/ijms13021598
Thippeswamy G, Sheela ML, Salimath BP (2008) Octacosanol isolated from Tinospora cordifolia downregulates VEGF gene expression by inhibiting nuclear translocation of NF-<kappa>B and its DNA binding activity. Eur J Pharmacol 588:141–150. https://doi.org/10.1016/j.ejphar.2008.04.027
Deacon CF (2007) Dipeptidyl peptidase 4 inhibition with sitagliptin: a new therapy for Type 2 diabetes. Expert Opin Investig Drugs 16:533–545. https://doi.org/10.1517/13543784.16.4.533
Wang C, Cai Z, Wang W et al (2020) Piperine regulates glycogen synthase kinase-3β-related signaling and attenuates cognitive decline in d-galactose-induced aging mouse model. J Nutr Biochem 75:108261. https://doi.org/10.1016/j.jnutbio.2019.108261
Singh SS, Pandey SC, Srivastava S et al (2003) Chemistry and medicinal properties of Tinospora cordifolia (Guduchi). Indian J Pharmacol 35:83–91
Xie J, Wu J, Yang S, Zhou H (2021) Network pharmacology-based study on the mechanism of Aloe vera for treating cancer. Evid Based Complement Altern Med 2021:1–8. https://doi.org/10.1155/2021/6077698
Dilman VM, Revskoy SY, Golubev AG (1986) Neuroendocrine-ontogenetic mechanism of aging: toward an integrated theory of aging. In: International review of neurobiology. Elsevier, Amsterdam, pp 89–156
Frolkis VV (1988) A hundred questions on neurohumoral mechanisms of aging. Gerontology 34:6–13. https://doi.org/10.1159/000212924
Yanai H, Budovsky A, Barzilay T et al (2017) Wide-scale comparative analysis of longevity genes and interventions. Aging Cell 16:1267–1275. https://doi.org/10.1111/acel.12659
Wang JC, Bennett M (2012) Aging and atherosclerosis: mechanisms, functional consequences, and potential therapeutics for cellular senescence. Circ Res 111:245–259. https://doi.org/10.1161/CIRCRESAHA.111.261388
Di Benedetto G, Iannucci LF, Surdo NC et al (2021) Compartmentalized signaling in aging and neurodegeneration. Cells 10:464. https://doi.org/10.3390/cells10020464
Chandran R, Kumar M, Kesavan L et al (2019) Cellular calcium signaling in the aging brain. J Chem Neuroanat 95:95–114. https://doi.org/10.1016/j.jchemneu.2017.11.008
Aunan JR, Cho WC, Søreide K (2017) The biology of aging and cancer: a brief overview of shared and divergent molecular hallmarks. Aging Dis 8:628. https://doi.org/10.14336/AD.2017.0103
Ryan AS (2000) Insulin resistance with aging: effects of diet and exercise. Sports Med 30:327–346. https://doi.org/10.2165/00007256-200030050-00002
Yeo E-J (2019) Hypoxia and aging. Exp Mol Med 51:1–15. https://doi.org/10.1038/s12276-019-0233-3
Gao Q, Tian D, Han Z et al (2021) Network pharmacology and molecular docking analysis on molecular targets and mechanisms of Buyang Huanwu decoction in the treatment of ischemic stroke. Evid Based Complement Altern Med 2021:1–15. https://doi.org/10.1155/2021/8815447
Acknowledgements
The authors would like to thank the Head of the Botany Department, Soban Singh Jeena University, S.S.J. Campus, Almora (Uttarakhand), India, for providing the facility and space for this research work. The authors would also like to acknowledge the Department of Botany, Kumaon Kesari Pt. Badridutt Pandey Government Post Graduate College, Bageshwar, Soban Singh Jeena University, Almora, for providing external support.
Funding
There was no funding source to carry out this research work.
Author information
Authors and Affiliations
Contributions
AB wrote the manuscript and done all the experiments parts. DT analyzed and interpreted the data. SC conceptualized and designed the work. SK and SC supervised the study. The whole manuscript was approved by all authors.
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare to have no actual or potential conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bisht, A., Tewari, D., Kumar, S. et al. Network pharmacology, molecular docking, and molecular dynamics simulation to elucidate the mechanism of anti-aging action of Tinospora cordifolia. Mol Divers (2023). https://doi.org/10.1007/s11030-023-10684-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11030-023-10684-w