Abstract
Background
Offspring consistently exhibit similar imaging features as their parents in cases of degenerative lumbar scoliosis (DLS). Nevertheless, the role of genetic factors in the pathogenesis of DLS remains uncertain.
Methods
A prospective analysis was conducted on 35 patients with DLS and their 36 offspring. Genomic DNA was extracted from 71 blood samples for gene mutation analysis using whole exome sequencin. Various demographic and imaging parameters were compared.
Results
In 11 pedigrees of the 35 family members with DLS, 13 suspected pathogenic genes were identified. Among the 35 DLS patients, 11/35(31.5%) exhibited susceptibility gene mutations (mutant group), while 24/35(68.5%) had no pathogenic gene mutations (non-mutant group). AVR was more severe in mutant group than that in no-mutant group (p < 0.05). Among the 36 offspring, 11/36(30.6%) cohorts presented susceptibility genes (mutant group), 25/36(69.4%) cohorts presented no pathogenic genes (no-mutant group). More cohorts in the mutant group presented vertebral rotation (72.8%) and scoliosis (45.5%) than those (24%), (12%) in the no-mutant group, respectively (p < 0.05). Among the 36 offspring, 8/36(22.2%) presented scoliosis (study group), they all presented the same scoliosis orientation and apex vertebrae/disc location to their parents, the other 28/36(77.8%) cohorts without scoliosis were enrolled as control group, the mutation rate (62.5%) was higher in study group than that (21.4%) in control group.
Conclusions
Genetic influences are significant in the onset of DLS, with affected families showing similar scoliosis patterns and identical apex vertebrae. Moreover, individuals with genetic mutations tend to have more pronounced vertebral rotation and at a higher risk of developing scoliosis.
Similar content being viewed by others
Introduction
The etiology of degenerative lumbar scoliosis (DLS) is multifactorial, mainly including genetic predisposition and environmental factors [1, 2]. Recent studies suggest that specific gene variants are associated with an increased likelihood of developing spinal deformities, including scoliosis [3, 4]. These genetic markers serve as potential indicators for early diagnosis and risk assessment. One of the pivotal genes identified is the GPR126 gene, which is involved in the regulation of intervertebral disc development, variants in this gene have been shown to affect the structural integrity of the spine, leading to increased susceptibility to degeneration and subsequent curvature [5]. Additionally, the collagen gene COL11A1 is vital for the structural integrity of ligaments and discs, implicating its role in the development of DLS [6].
The current therapeutic strategies for DLS focus on symptom management and improving quality of life. Physical therapy, bracing, and pain management through medications are commonly employed non-surgical methods [7]. Theoretically, the development of gene therapy techniques that can modify the expression of specific genes associated with scoliosis holds promise for future treatments [8]. Additionally, nutritional and lifestyle interventions tailored to an individual’s genetic makeup could play a significant role in delaying the onset or progression of the disease. It is also worth considering the integration of genetic screening in routine clinical practice to identify at-risk individuals and provide early intervention strategies. It becomes increasingly clear that a personalized approach, grounded in the genetic profile of each patient, could revolutionize the standard of care for those affected by this debilitating condition.
Nevertheless, there is currently a lack of evidence from family studies, the pathogenic genes understanding of DLS remains poor. In the current study, we present strategies for whole exome sequencing studies to detect point mutation, deletion and insertion underlying DLS susceptibility and aim to explore the effects of mutations in pathogenic genes.
Materials and methods
Clinical samples
This prospective study was approved by the Institutional Review Board (IRB) of the Third Hospital of HeBei Medical University (S2022-094-1) in accordance with the Declaration of Helsinki. Before data collection and analysis, each patient provided informed consent and use of blood samples has been agreed by all individuals. The Inclusion criteria included (1) DLS patients with age older than 50 years, (2) The offspring (son or daughter) was ≥ 35 years old, and had lumbar Postero-anterior and lateral X-rays, (3) The patient and offspring were both enrolled. Exclusion criteria were (1) previous history of lumbar surgery, (2) bone tumor, or systemic chronic diseases (ankylosing spondylitis, rheumatoid arthritis, pyogenic spondylitis, or tuberculosis, etc.), which may potentially affect paravertebral muscle atrophy, (3) patients with missing imaging data or the anatomical identification was difficult to recognize for radiological measurement. Thirty-five consecutive families including 35 DLS patients and their offspring (36 cohorts) participated in the study during the period from June 2022 to December 2023.
X-ray assessment and MRI measurement
Bone structure parameters, including Cobb angle, apical vertebrae location (AVL), apical vertebral translation (AVT), apical vertebral rotation (AVR) based on Nash-Moe method [12], lumbar lordosis (LL) and sacral slope (SS, ) were measured from neutral standing Postero-anterior and lateral radiographs. The lumbar spine was examined using a 3.0 T MR scanner. T2-weighted (T2W) images were obtained with TR/TE times of 3200.00 ms/108.00 ms and a slice thickness of 4 mm. The center slice of the Images corresponding to the apical vertebral was chosen for the paraspinal muscle (PM) evaluation. The cross-sectional areas (CSAs) of the multifidus and erector spine muscles at apex of main curve were measured by outlining the fascial boundary of the muscle using the Image J - win64 software. The fat infiltration area (FIA) in the total CSA of the muscle was evaluated using a threshold technique. The fat infiltration rate (FIR) was calculated as the percentage of FIA to the total CSAs (Fig. 1). Each data was assessed 3 times, and the average value was calculated as the final data.
Extraction of DNA and whole exome sequencing (WES)
A blood sample (2 mL) was collected from each subject in ethylenediaminetetraacetic acid (EDTA) tube for the genomic DNA extraction. Genomic DNA was extracted with 250 µL whole blood using DNA isolation kit for mammalian blood (Magen, D3111, China) and stored at -20 °C before use. The primers, probes and reaction conditions are available on request. The WES was examined using next generation sequencing (NGS; MGI, DNBSEQ-T7) and the number of target genes is 19,500. The length of the target region is 41,888,254 bp, and the coverage of the target area is 99.98%. The average depth of target area is 164.53X, and the mean target coverage (> 20X) was 99.7%. All mutation sites were detected and confirmed using Sanger sequencing (ABI 3730XL).
Statistical analysis
Data were analyzed using Statistical Product and Service Solutions software (version 17; SPSS, Chicago, IL). Three-way Cohen’s Kappa correlation coefficients were calculated to assess the intra- and interobserver reliability for measuring the radiological data. Continuous variables were recorded as mean ± standard deviation, and categorical variables were expressed as frequency or percentages. An independent t test was used to analyze the difference of continuous variables. An χ2 analysis and Fisher’s exact test were used to examine the differences among categorical variables. The statistical significance was set at p < 0.05.
Results
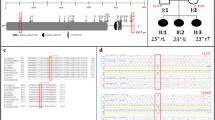
All the DNA samples were greater than 10 kb in molecular weight, which confirms intactness of the genome. Neither DNA degradation nor RNA contamination was observed (Fig. 2). The whole exome sequencing was examined for each sample, and mutation point was verified by Sanger sequencing analysis. In the 35 family, 13 suspected pathogenic genes were discovered in 11 pedigrees, which include 11 DLS patients and 11 offspring, details see Table 1. Some of them are classified in ClinVar as missense variants with uncertain significance (ACAN c.6838G > A; FGFR3 c.1106 C > T; COL1A2 c.2024G > A; FLNB c.2484 + 5G > A; RYR1 c.12473G > A; FBN2 c.139G > T). Only the variant MAPK7 c.886G > A is classified as pathogenic (no assertion criteria provided). Other genetic variants including COL5A2 c.529G > A; COL12A1 c.7463 C > A; FLNA c.5425 C > T; FN1 c.4516 C > T; POC5 c.1439 A > G; FBN1 c.4449G > A were not included in the ClinVar database. Most notably, this is the first study to our knowledge to report those susceptibility genes (other genetic variants) and these missense variants including COL1A2 c.2024G > A; FLNB c.2484 + 5G > A; RYR1 c.12473G > A; FBN2 c.139G > T. For example, the family #3 in Table 1. Exome sequencing identified a novel heterozygous variant in exon 47 of the COL12A1 gene: NM_004370.5: c.7463 C > A, (p. Ser2488Tyr) and the COL12A1 variant (c.7463 C > A) was confirmed by Sanger sequencing in the proband’s family (Figs. 3 and 4).
Among the 35 DLS patients, 11/35(31.5%) patients presented susceptibility genes and enrolled as mutant group, 24/35(68.5%) patients presented no pathogenic genes and enrolled as no-mutant group. AVR was more severe in mutant group than that in no-mutant group (X2 = 6.258, p = 0.030), there was no significant difference in age, gender, BMI, scoliosis orientation, Cobb angle, vertebrae number in main curve, AVT, AVL, SS, LL, FIR of paraspinal muscle between mutant and no-mutant group in the 35 DLS patients (Table 3).
Among the 36 offspring, 11/36(30.6%) cohorts presented candidate-genes and enrolled as mutant group, 25/36(69.4%) cohorts presented no pathogenic genes and enrolled as no-mutant group. More cohorts in the mutant group presented AVR (X2 = 5.719, p = 0.017) and scoliosis (X2 = 4.946, p = 0.026) than those in the no-mutant group. There was no significant difference in age, gender, BMI between mutant and no-mutant group in the 36 offspring (Table 4).
Among the 36 offspring, 8/36(22.2%) presented scoliosis based on diagnosis criteria of 10 degrees or above, these cohorts were enrolled as study group, they all presented the same scoliosis orientation and apex vertebrae/disc location with their parents (Table 2; Fig. 5). The other 28/36(77.8%) cohorts without lumbar coronal misalignment (Cobb’s angle was smaller than 10 degrees) were enrolled as control group, the mutation rate was higher in study group than that in control group, there was no significant difference in age, gender, BMI, SS, LL between the two cohorts. (Table 5)
Discussion
It is noteworthy that out of the 35 patients with scoliosis 68.5% had normal genes and only 31.5% had gene mutations, and that AVR was more severe in mutant cohorts than that in no-mutant cohorts both in DLS patients and their offspring. Additionally, more scoliosis was detected in offspring cohorts with susceptibility genes, they all presented the same scoliosis orientation and apex vertebrae/disc location with their parents. Our research delved into the possibility of a genetic predisposition to the specific scoliosis phenotype, which could be attributed to the inheritance of pathogenic genes. We hypothesized that these genes might influence skeletal development and spinal alignment through pathways that are not yet fully understood. To explore this hypothesis, we conducted a comprehensive analysis of the genomic data from our patient cohorts and denote that variants such as COL12A1, FLNA, FLNB, FN1, MAPK7, RYR1, POC5, FBN1, and FBN2 may exhibit associations with DLS phenotypes. It is imperative to underscore that the presence of DLS-associated variants does not establish causality but rather signifies potential candidate genes in linkage disequilibrium with the disease. Intriguingly, these genes were found to be involved in the regulation of bone morphogenesis, extracellular matrix organization, and signaling pathways that have been implicated in vertebral development. This suggests that mutations in these genes could disrupt normal spinal development processes, leading to the manifestation of a similar scoliosis phenotype in affected family members.
Collagen Type XII, encoded by the COL12A1 gene, is integral to the extracellular matrix’s architecture through its interaction with other matrix macromolecules [9]. This interaction is critical for preserving the structural integrity of connective tissue. Pathogenic variants in COL12A1 can perturb these molecular interactions, leading to compromised assembly of the fibronectin-collagen matrix, which is indispensable for the biomechanical resilience and pliability of spinal tissue [10]. Such disturbances may induce alterations in the biomechanical properties of the vertebral column. Cells harboring these mutations demonstrate aberrant mechanotransduction signaling, a process essential for cellular adaptation to mechanical stimuli [11]. This dysregulated signaling is implicated in the etiology of abnormal spinal curvature due to the inability to adequately redistribute mechanical forces, potentially exacerbating the progression of scoliosis. Given these insights, we advocate for the incorporation of COL12A1 mutation screening in clinical settings to preemptively identify individuals predisposed to DLS. Additionally, the development of pharmacotherapies aimed at the molecular derangements observed in patients with COL12A1 mutations holds promise. Therapeutic agents that fortify the extracellular matrix or modulate mechanotransduction pathways may offer clinical benefits.
The FLNA and FLNB loci are responsible for the synthesis of filamin A and B, which are integral cytoplasmic actin-binding proteins that are pivotal in sustaining cellular architecture and modulating intracellular signaling cascades [12]. These proteins influence the architectural integrity of the cytoskeleton, cellular adherence, and the modulation of signaling networks. Mutations within these genes may perturb these critical processes, potentially altering the biomechanical attributes of the soft connective tissue within the lumbar spine, thereby contributing to the pathogenesis of scoliotic deviations [13]. Specifically, the resultant aberrant protein may impair the biomechanical functionality of the intervertebral discs and ligaments, heightening their vulnerability to degeneration when subjected to normal physiological loads. The ensuing degradation of these connective structures can lead to disproportionate mechanical stress on the vertebral column, which could amplify spinal curvature, culminating in scoliosis. The incorporation of genetic assays for FLNA and FLNB mutations into the diagnostic algorithm for scoliosis could enable precocious clinical interventions, potentially decelerating the disease’s progression. Moreover, longitudinal investigations tracking individuals harboring FLNA and FLNB mutations could illuminate the etiopathogenic trajectory of DLS and pinpoint opportune junctures for clinical intervention.
The FN1 locus, responsible for the synthesis of fibronectin, is integral to cellular adhesion, proliferation, motility, and tissue repair processes [14]. Genetic aberrations within this locus may disrupt the mechanical attributes of the lumbar spine’s extracellular matrix, culminating in asymmetric mechanical stress distribution and ensuing spinal malformation. Prior investigations have established the prevalence of fibronectin within the nucleus pulposus of intervertebral discs, underscoring its necessity for disc integrity [15]. An allelic variation in FN1 may undermine the structural and functional coherence of the intervertebral disc, potentially instigating or aggravating degenerative disc disease.
The allelic variation within the MAPK7 gene, which influences the enzymatic activity of mitogen-activated protein kinase 7, plays a crucial role in the MAPK signal transduction cascade [16]. This cascade is involved in an array of cellular mechanisms such as cell growth, morphological differentiation, and programmed cell death. Mutations in the MAPK7 locus may perturb these integral vertebral processes, potentially precipitating anomalous spinal curvature formation [17]. Theoretically, a polymorphism in MAPK7 could compromise the viability and function of bone-forming osteoblasts or bone-resorbing osteoclasts, thereby altering skeletal homeostasis and possibly exacerbating the development of scoliosis.
The RYR1 locus is responsible for coding the ryanodine receptor, an integral transmembrane channel facilitating the efflux of calcium ions from the sarcoplasmic reticulum within myocytes [18]. Pathogenic variants in this locus can lead to dysregulation of calcium ionostasis, potentially impairing myofibrillar contraction dynamics and sequentially influencing axial skeleton biomechanics [19]. Electrodiagnostic studies, in the form of electromyography, alongside histopathological examination of muscular tissue, can elucidate the pathophysiological impact of RYR1 aberrations on musculoskeletal function and their role in the pathogenesis of progressive lumbar spinal deformities.
The POC5 gene, known for its involvement in centriole formation, may influence the integrity of the cytoskeletal structure within spinal cells, thereby affecting their ability to maintain proper alignment and resist degenerative forces [20]. Recent studies have suggested that the aberrant expression of POC5 can lead to altered microtubule dynamics, which, in turn, could compromise the structural stability of intervertebral cells [21]. This instability may predispose the spinal column to abnormal curvature, particularly under the continuous mechanical stress experienced by the lumbar region. Furthermore, the presence of the POC5 variant has been correlated with an accelerated degeneration of disc tissue, exacerbating the propensity for scoliosis development.
The FBN1 gene encodes fibrillin-1, a glycoprotein that is essential for the formation of elastic fibers within connective tissue [22]. Mutations in this gene have been associated with Marfan syndrome, which features a range of skeletal abnormalities. The FBN2 gene, on the other hand, encodes fibrillin-2, playing a similar, yet distinct role in the microfibrillar network, primarily during early development [23]. Further exploration of the pathophysiology reveals that aberrations in these genes could disrupt the intricate balance of extracellular matrix production and degradation, leading to compromised structural integrity of the spinal column. This imbalance may result in the vertebrae becoming susceptible to asymmetric loading and the subsequent development of scoliosis.
In conclusion, our findings emphasize the significance of genetic factors in the development and progression of scoliosis. The identification of susceptibility genes and their associated pathways offers valuable insights into the etiology of this condition and presents new opportunities for early diagnosis and intervention. Future research should focus on the functional characterization of these genes to unravel the precise mechanisms by which they contribute to scoliosis pathogenesis. This could lead to the development of novel therapeutic approaches that target the molecular basis of the disease, offering hope for improved patient outcomes. There were several potential limitations in this study. First, only Chinese Han individuals were included in the current study and Ethnic variation was not covered. Second, the number of patients included was relatively small, and the study may be under powered to detect the potential susceptibility genes that may influenced the DLS, further validation of large sample clinical data from multiple centers is needed in the future.
Data availability
No datasets were generated or analysed during the current study.
References
Shin JH, Ha KY, Jung SH, et al. Genetic predisposition in degenerative lumbar scoliosis due to the copy number variation. Spine. 2011;36:1782–93. https://doi.org/10.1097/BRS.0b013e318221a65f.
Park YS, Suh KT, Shin JK, et al. Estrogen receptor gene polymorphism in patients with degenerative lumbar scoliosis. Br J Neurosurg. 2017;31:63–6. https://doi.org/10.1080/02688697.2016.1206186.
Shi X, Li P, Wu X, et al. Whole-transcriptome sequencing identifies key differentially expressed circRNAs/lncRNAs/miRNAs/mRNAs and linked ceRNA networks in adult degenerative scoliosis. Front Mol Neurosci. 2023;16:1038816. https://doi.org/10.3389/fnmol.2023.1038816.
Shi X, Li P, Wu X, et al. RNA-Seq Comprehensive Analysis reveals the long noncoding RNA expression Profile and Coexpressed mRNA in adult degenerative scoliosis. Front Genet. 2022;13:902943. https://doi.org/10.3389/fgene.2022.902943.
Akbik OS, Ban VS, MacAllister MC, et al. Genetic and serum markers in adult degenerative scoliosis: a literature review. Spine Deform. 2022;10:479–88. https://doi.org/10.1007/s43390-021-00451-y.
Jiang H, Yang Q, Jiang J, et al. Association between COL11A1 (rs1337185) and ADAMTS5 (rs162509) gene polymorphisms and lumbar spine pathologies in Chinese Han population: an observational study. BMJ Open. 2017;7:e015644. https://doi.org/10.1136/bmjopen-2016-015644.
Lai J, Tan H, Feng H, et al. Treatment of degenerative lumbar scoliosis using transforaminal lumbar interbody fusion based on the concept of intervertebral correction. Int Orthop. 2023;47:1303–13. https://doi.org/10.1007/s00264-023-05774-1.
Sivakamasundari V, Lufkin T. Stemming the degeneration: IVD stem cells and stem cell regenerative therapy for degenerative disc disease. Adv Stem Cells. 2013;2013:724547. https://doi.org/10.5171/2013.724547.
Furuhata-Yoshimura M, Yamaguchi T, Izu Y, et al. Homozygous splice site variant affecting the first von Willebrand factor A domain of COL12A1 in a patient with myopathic Ehlers-Danlos syndrome. Am J Med Genet A. 2023;191:2631–9. https://doi.org/10.1002/ajmg.a.63328.
Shalmiev A, Nadeau G, Aaron M, et al. Genetic factors contributing to late adverse musculoskeletal effects in childhood acute lymphoblastic leukemia survivors. Pharmacogenomics J. 2022;22:19–24. https://doi.org/10.1038/s41397-021-00252-6.
van den Akker GG, Surtel DA, Cremers A, et al. Novel Immortal Cell lines support Cellular Heterogeneity in the human Annulus Fibrosus. PLoS ONE. 2016;11:e0144497. https://doi.org/10.1371/journal.pone.0144497.
Mizuhashi K, Kanamoto T, Moriishi T, et al. Filamin-interacting proteins, Cfm1 and Cfm2, are essential for the formation of cartilaginous skeletal elements. Hum Mol Genet. 2014;23:2953–67. https://doi.org/10.1093/hmg/ddu007.
Hu J, Lu J, Goyal A, et al. Opposing FlnA and FlnB interactions regulate RhoA activation in guiding dynamic actin stress fiber formation and cell spreading. Hum Mol Genet. 2017;26:1294–304. https://doi.org/10.1093/hmg/ddx047.
Shen N, Chen N, Zhou X, et al. Alterations of the gut microbiome and plasma proteome in Chinese patients with adolescent idiopathic scoliosis. Bone. 2019;120:364–70. https://doi.org/10.1016/j.bone.2018.11.017.
Cherif H, Mannarino M, Pacis AS, et al. Single-cell RNA-Seq analysis of cells from degenerating and non-degenerating intervertebral discs from the same individual reveals new biomarkers for intervertebral disc degeneration. Int J Mol Sci. 2022;23:3993. https://doi.org/10.3390/ijms23073993.
Wu C, Liu H, Zhong D, et al. Mapk7 deletion in chondrocytes causes vertebral defects by reducing MEF2C/PTEN/AKT signaling. Genes Dis. 2023;11:964–77. https://doi.org/10.1016/j.gendis.2023.02.012.
Zhou T, Chen C, Xu C, et al. Mutant MAPK7-Induced Idiopathic Scoliosis is linked to impaired Osteogenesis. Cell Physiol Biochem. 2018;48:880–90. https://doi.org/10.1159/000491956.
AlBakri A, Karaoui M, Alkuraya FS, et al. Congenital ptosis, scoliosis, and malignant hyperthermia susceptibility in siblings with recessive RYR1 mutations. J AAPOS. 2015;19:577–9. https://doi.org/10.1016/j.jaapos.2015.08.006.
Shaaban S, Ramos-Platt L, Gilles FH, et al. RYR1 mutations as a cause of ophthalmoplegia, facial weakness, and malignant hyperthermia. JAMA Ophthalmol. 2013;131:1532–40. https://doi.org/10.1001/jamaophthalmol.2013.4392.
Hassan A, Parent S, Mathieu H, et al. Adolescent idiopathic scoliosis associated POC5 mutation impairs cell cycle, cilia length and centrosome protein interactions. PLoS ONE. 2019;14:e0213269. https://doi.org/10.1371/journal.pone.0213269.
Xu L, Sheng F, Xia C, et al. Common variant of POC5 is Associated with the susceptibility of adolescent idiopathic scoliosis. Spine. 2018;43:E683–8. https://doi.org/10.1097/BRS.0000000000002490.
de Azevedo GBL, Perini JA, Araújo Junior AEP, et al. Association of FBN1 polymorphism with susceptibility of adolescent idiopathic scoliosis: a case-control study. BMC Musculoskelet Disord. 2022;23:430. https://doi.org/10.1186/s12891-022-05370-1.
Sheng F, Xia C, Xu L, et al. New evidence supporting the role of FBN1 in the development of adolescent idiopathic scoliosis. Spine. 2019;44:E225–32. https://doi.org/10.1097/BRS.0000000000002809.
Acknowledgements
None. The manuscript submitted does not contain information about medical device(s)/drug(s).
Funding
Natural Science Foundation of Hebei Province, H2022206056.
Author information
Authors and Affiliations
Contributions
ZGS was responsible for interpreting results and writing this article. YLL and HRY was responsible for data collection, statistics analysis. YCH and DZ was responsible for English editing. WSL and WYD was responsible for English editing. HW was responsible for designing the search strategy, evaluating the articles and English editing.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the institutional review board of the Third Hospital of HeBei Medical University. Informed consent to participate was obtained from all of the participants in the study.
Human Ethics and Consent to Participate
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Shang, Z., Liu, Y., Yuan, H. et al. Inherited genetic predisposition and imaging concordance in degenerative lumbar scoliosis patients and their descendants. J Orthop Surg Res 19, 494 (2024). https://doi.org/10.1186/s13018-024-05000-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13018-024-05000-7