Abstract
Study Design
A hypothesis-driven study was conducted in a familial cohort to determine the potential association between variants within the T-box 6 (TBX6) gene and familial idiopathic scoliosis (FIS).
Objective
To determine whether variants within exons of the TBX6 gene segregate with the FIS phenotype within a sample of families with FIS.
Summary of Background Data
Idiopathic scoliosis is a structural curvature of the spine whose underlying genetic etiology has not been established. Idiopathic scoliosis has been reported to occur at a higher rate than expected in family members of individuals with congenital scoliosis, which suggests that the 2 diseases might have a shared etiology. The TBX6 gene on chromosome 16p, essential to somite development, has been associated with congenital scoliosis in a Chinese population. Previous studies have identified linkage to this locus in families with FIS, and specifically with rs8060511, located in an intron of the TBX6 gene.
Methods
Parent-offspring trios from 11 families (13 trios; 42 individuals) with FIS were selected for Sanger sequencing of the TBX6 gene. Trios were selected from a large population of families with FIS in which a genome-wide scan had resulted in linkage to 16p.
Results
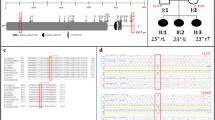
Sequencing analyses of the subset of families resulted in the identification of 5 coding variants. Three of the five variants were novel; the remaining 2 variants had previously been characterized and they account for 90% of the observed variants in these trios. In all cases, there was no correlation between transmission of the TBX6 variant allele and FIS phenotype. However, an analysis of regulatory markers in osteoblasts showed that rs8060511 is in a putative enhancer element.
Conclusions
Although this study did not identify any TBX6 coding variants that segregate with FIS, we identified a variant that is located in a potential TBX6 enhancer element. Therefore, further investigation of the region is needed.
Similar content being viewed by others
References
Bonaiti C, Feingold J, Briard ML, et al. Genetics of idiopathic scoliosis. Helv PaediatrActa 1976;31:229–40.
Cowell HR, Hall JN, MacEwen GD. Genetic aspects of idiopathic scoliosis: a Nicholas Andry Award essay, 1970. Clin Orthop Relat Res 1972;86:121–31.
Czeizel A, Bellyei A, Barta O, Magda T, Molnar L. Genetics of adolescent idiopathic scoliosis. J Med Genet 1978;15:424–7.
Risehorough EJ, Wynne-Davies R. A genetic survey of idiopathic scoliosis in Boston, Massachusetts. J Bone Joint Surg Am 1973;55:974–82.
Miller NH, Marosy B, Justice CM, et al. Linkage analysis of genetic loci for kyphoscoliosis on chromosomes 5p13, 13q13.3, and 13q32. Am J Med Genet A 2006;140:1059–68.
Gao X, Gordon D, Zhang D, et al. Chd7 gene polymorphisms are associated with susceptibility to idiopathic scoliosis. Am J Hum Genet 2007;80:957–65.
Kou I, Takahashi Y, Johnson TA, et al. Genetic variants in gprl26 are associated with adolescent idiopathic scoliosis. Nat Genet 2013;45:676–9.
Sharma S, Gao X, Londono D, et al. Genome-wide association studies of adolescent idiopathic scoliosis suggest candidate susceptibility genes. Hum Mol Genet 2011;20:1456–66.
Takahashi Y, Kou I, Takahashi A, et al. A genome-wide association study identifies common variants near lbxl associated with adolescent idiopathic scoliosis. Nat Genet 2011;43:1237–40.
Wardle FC, Papaioannou VE. Teasing out t-box targets in early mesoderm. Curr Opin Genet Dev 2008;18:418–25.
Giampietro PF, Blank RD, Raggio CL, et al. Congenital and idiopathic scoliosis: clinical and genetic aspects. Clin Med Res 2003;1:125–36.
Purkiss SB, Driscoll B, Cole WG, Alman B. Idiopathic scoliosis in families of children with congenital scoliosis. Clin Orthop Relat Res 2002;401:27–31.
McKinley LM, Leatherman KD. Idiopathic and congenital scoliosis in twins. Spine (Phila Pa 1976) 1978;3:227–9.
Miller NH, Justice CM, Marosy B, et al. Identification of candidate regions for familial idiopathic scoliosis. Spine (Phila Pa 1976) 2005;30:1181–7.
Axenovich TI, Zaidman AM, Zorkoltseva IV, et al. Segregation analysis of idiopathic scoliosis: demonstration of a major gene effect. Am J Med Genet 1999;86:389–94.
Fei Q, Wu Z, Wang H, Zhou X, et al. The association analysis of tbx6 polymorphism with susceptibility to congenital scoliosis in a Chinese Han population. Spine (Phila Pa 1976) 2010;35:983–8.
Sparrow DB, Mclnerney-Leo A, Gucev ZS, et al. Autosomal dominant spondylocostal dysostosis is caused by mutation in tbx6. Hum Mol Genet 2013;22:1625–31.
Miller NH, Justice CM, Marosy B, et al. Intra-familial tests of association between familial idiopathic scoliosis and linked regions on 9q31.3-q34.3 and 16pl2.3-q22.2. Hum Hered 2012;74:36–44.
McCarthy SE, Makarov V, Kirov G, et al. Microduplications of 16p11.2 are associated with schizophrenia. Nat Genet 2009;41:1223–7.
Zufferey F, Sherr EH, Beckmann ND, et al. A 600 kb deletion syndrome at 16p11.2 leads to energy imbalance and neuropsychiatric disorders. J Med Genet 2012;49:660–8.
Al-Kateb H, Khanna G, Filges I, et al. Scoliosis and vertebral anomalies: additional abnormal phenotypes associated with chromosome 16p11.2 rearrangement. Am J Med Genet A 2014;164:1118–26.
Shimojima K, Inoue T, Fujii Y, et al. A familial 593-kb microdele-tion of 16p11.2 associated with mental retardation and hemiverte-brae. Eur J Med Genet 2009;52:433–5.
Justice CM, Miller NH, Marosy B, et al. Familial idiopathic scoliosis: evidence of an x-linked susceptibility locus. Spine (Phila Pa 1976) 2003;28:589–94.
Armstrong GW, Livermore III NB, Suzuki N, Armstrong JG Nonstandard vertebral rotation in scoliosis screening patients: its prevalence and relation to the clinical deformity. Spine (Phila Pa 1976) 1982;7:50–4.
Kane WJ. Scoliosis prevalence: a call for a statement of terms. Clin Orthop Relat Res 1977;126:43–6.
Shands Jr AR, Eisberg HB. The incidence of scoliosis in the state of delaware; a study of 50,000 minifilms of the chest made during a survey for tuberculosis. J Bone Joint Surg Am 1955;37:1243–9.
Sambrook J, Fritsch EF, Maniatis T Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989.
Moore D, Dowhan D. Purification and Concentration of DNA from Aqueous Solutions. CurrProtoc Molec Biol 2002;59:I:2.1A:2.1–2.1.10.
Comings DE, Gade-Andavolu R, Gonzalez N, et al. Multivariate analysis of associations of 42 genes in adhd, odd and conduct disorder. Clin Genet 2000;58:31–40.
Rabinowitz D, Laird N. A unified approach to adjusting associa-tion tests for population admixture with arbitrary pedigree struc-ture and arbitrary missing marker information. Hum Hered 2000;50:211–23.
Horvath S, Xu X, Lake SL, et al. Family-based tests for associating haplotypes with general phenotype data: application to asthma genetics. Genet Epidemiol 2004;26:61–9.
Laird NM, Horvath S, Xu X. Implementing a unified approach to family-based tests of association. Genet Epidemiol 2000; 19(Suppl 1):S36–42.
Barrett JC, Fry B, Mailer J, Daly MJ. Haploview: analysis and visualization of Id and haplotype maps. Bioinformatics 2005;21:263–5.
Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at USCS. Genome Res 2002;12:996–1006.
Karolchik D, Barber GP, Casper J, et al. The ucsc genome browser database: 2014 update. Nucleic Acids Res 2014;42:D764–70.
Karolchik D, Hinrichs AS, Furey TS, et al. The ucsc table browser data retrieval tool. Nucleic Acids Res 2004;32:D493–6.
ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74.
Zhou X, Maricque B, Xie M, et al. The human epigenome browser at Washington University. Nat Methods 2011;8:989–90.
Zhou X, Wang T Using the Wash U Epigenome Browser to Examine Genome-Wide Sequencing Data. Curr Protoc Bioinformatics 2012;40:10.10:10.10.1–10.10.14.
Shinawi M, Liu P, Kang SH, et al. Recurrent reciprocal 16p11.2 rearrangements associated with global developmental delay, behavioural problems, dysmorphism, epilepsy, and abnormal head size. J Med Genet 2010;47:332–41.
Bijlsma EK, Gijsbers AC, Schuurs-Hoeijmakers JH, et al. Extending the phenotype of recurrent rearrangements of 16p11.2: deletions in mentally retarded patients without autism and in normal individuals. Eur J Med Genet 2009;52:77–87.
Fernandez BA, Roberts W, Chung B, et al. Phenotypic spectrum associated with de novo and inherited deletions and duplications at 16p11.2 in individuals ascertained for diagnosis of autism spectrum disorder. J Med Genet 2010;47:195–203.
Schaaf CP, Goin-Kochel RP, Nowell KP, et al. Expanding the clinical spectrum of the 16p11.2 chromosomal rearrangements: three patients with syringomyelia. Eur J Hum Genet 2011;19:152–6.
Shen Y, Chen X, Wang L, et al. Intra-family phenotypic heterogeneity of 16p11.2 deletion carriers in a three-generation Chinese family. Am J Med Genet B Neuropsychiatr Genet 2011;156:225–32.
Smit, AFA, Hubley, R & Green, P. RepeatMasker Open-3.0. 1996–2010. http://www.repeatmasker.org.
Di Benedetto A, Watkins M, Grimston S, et al. N-cadherin and cadherin 11 modulate postnatal bone growth and osteoblast differentiation by distinct mechanisms. J Cell Sci 2010;123:2640–8.
Griffin KJ, Kimelman D. One-eyed pinhead and spadetail are essential for heart and somite formation. Nat Cell Biol 2002;4:821–5.
Nakaya MA, Biris K, Tsukiyama T, et al. Wnt3a links left-right determination with segmentation and anteroposterior axis elongation. Development 2005;132:5425–36.
Yamaguchi TP, Takada S, Yoshikawa Y, et al. T (brachyury) is a direct target of wnt3a during paraxial mesoderm specification. Genes Dev 1999;13:3185–90.
Showell C, Binder O, Conlon FL. T-box genes in early embryogen-esis. Dev Dynm 2004;229:201–18.
Naiche LA, Harrelson Z, Kelly RG, Papaioannou VE. T-box genes in vertebrate development. Annu Rev Genet 2005;39:219–39.
Martin BL, Kimelman D. Regulation of canonical wnt signaling by brachyury is essential for posterior mesoderm formation. Dev Cell 2008;15:121–33.
Chapman DL, Cooper-Morgan A, Harrelson Z, Papaioannou VE. Critical role for tbx6 in mesoderm specification in the mouse embryo. Mech Dev 2003;120:837–47.
Ghebranious N, Blank RD, Raggio CL, et al. A missense t (bra-chyury) mutation contributes to vertebral malformations. J Bone Miner Res 2008;23:1576–83.
Chapman DL, Papaioannou VE. Three neural tubes in mouse embryos with mutations in the t-box gene tbx6. Nature 1998;391:695–7.
Watabe-Rudolph M, Schlautmann N, Papaioannou VE, Gossler A. The mouse rib-vertebrae mutation is a hypomorphic tbx6 allele. Mech Dev 2002;119:251–6.
Author information
Authors and Affiliations
Corresponding author
Additional information
Author disclosures: EEB (none); KS (none); CMJ (none); RMB (none); AP (none); CIW (none); AP (none); OP (none); OT (none); NHM (none).
Rights and permissions
About this article
Cite this article
Baschal, E.E., Swindle, K., Justice, C.M. et al. Sequencing of the TBX6 Gene in Families With Familial Idiopathic Scoliosis. Spine Deform 3, 288–296 (2015). https://doi.org/10.1016/j.jspd.2015.01.005
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1016/j.jspd.2015.01.005