Abstract
Background
Melanoma is the deadliest of skin cancers and has an increasing annual incidence worldwide. It is a multi-factorial disease most likely arising from both genetic predisposition and environmental exposure to ultraviolet light. Genetic variability of the components of the biological circadian clock is recognized to be a risk factor for different type of cancers. Moreover, two variants of a clock gene, RORA, have been associated with melanoma patient’s prognosis. Our aim is to test the hypothesis that specific single nucleotide polymorphisms (SNPs) of the circadian clock genes may significantly influence the predisposition to develop cutaneous melanoma or the outcome of melanoma patients.
Methods
We genotyped 1239 subjects, 629 cases of melanoma and 610 healthy controls in 14 known SNPs of seven selected clock genes: AANAT, CLOCK, NPAS2, PER1, PER2, RORA, and TIMELESS. Genotyping was conducted by q-PCR. Multivariate logistic regression was employed for susceptibility of melanoma assessment, modeled additively. Subgroup analysis was performed by gender. For the female subgroup, a further discrimination was performed by age. For prognosis of melanoma assessment, multivariate Cox proportional hazard regression was employed. The Benjamini–Hochberg method was utilized as adjustment for multiple comparisons.
Results
We identified two RORA SNPs statistically significant with respect to the association with melanoma susceptibility. Considering the putative role of RORA as a nuclear steroid hormone receptor, we conducted a subgroup analysis by gender. Interestingly, the RORA rs339972 C allele was associated with a decreased predisposition to develop melanoma only in the female subgroup (OR 0.67; 95% CI 0.51–0.88; P = 0.003) while RORA rs10519097 T allele was associated with a decreased predisposition to develop melanoma only in the male subgroup (OR 0.62; 95% CI 0.44–0.87; P = 0.005). Moreover, the RORA rs339972 C allele had a decreased susceptibility to develop melanoma only in females aged over 50 years old (OR 0.67; 95% CI 0.54–0.83; P = 0.0002). None of the studied SNPs were significantly associated with the prognosis.
Conclusions
Overall, we cannot ascertain that circadian pathway genetic variation is involved in melanoma susceptibility or prognosis. Nevertheless, we identified an interesting relationship between melanoma susceptibility and RORA polymorphisms acting in sex-specific manner and which is worth further future investigation.
Similar content being viewed by others
Background
Melanoma, which arises from the uncontrolled proliferation of melanocytes, is the deadliest of skin cancers and has an annual incidence of 22 new cases per 100,000 inhabitants in the US. It is a multi-factorial disease most likely arising from both genetic predisposition and environmental exposure to ultraviolet light. Phenotypical characteristics such as fair skin type, dysplastic nevi and multiple common nevi contribute to increase melanoma risk. In general, the predisposition to melanoma follows a polygenic model where in addition to the high penetrance genes with familial aggregation, as CDKN2A mutations, many low/medium penetrance genes are recognized with a risk-modifying role in the general population [1]. Approximately 5–10% of cutaneous melanoma cases occurs in a familial setting [2]. Genome-wide association studies (GWAS) and improved DNA sequencing techniques as next generation sequencing (NGS) contributed to elucidate the genetic basis of melanoma susceptibility with the identification of a plethora of new genetic loci associated with melanoma. Although, several genes responsible for melanoma predisposition have been identified, our knowledge on this field is still poorly understood [3].
An increasing number of epidemiological studies associates chronodisruption with the risk of developing various type of cancers, including melanoma [4, 5], leading the International Agency for Research on Cancer (IARC) to classify Shifwork (with night shifts, which involves circadian disruption) as a potential carcinogenic for humans (Group 2A) [6]. Regarding the relationship between the circadian clock machinery and melanoma, the majority of researchers focused on the expression of the circadian clock genes in melanoma cell lines and in skin biopsies in human or mouse. These studies showed that clock genes are downregulated in melanoma as compared to normal adjacent tissue (or compared to nevi) [7, 8]; moreover, the reduced clock gene expression is associated with increased tumor aggressiveness and a worse prognosis [7, 9]. Only one study addressed the issue on genetic variability of the promoter region of the NPAS2 clock gene and melanoma risk, and found a significant association with a polymorphic GGC repeat [10]. Another study examined the association of polymorphisms of steroid hormone receptors and melanoma prognosis, and found significant associations between cutaneous melanoma-specific survival and two SNPs on the clock gene RORA, whose putative protein encodes a ligand-activated transcription factor [11].
In our previous analysis on genetic predisposition to cutaneous melanoma, we studied the copy number variations (CNVs) of the transcription factor E2F1 [12] and found that 1.6% of melanoma patients harbored more than two copies of E2F1. The difference with the healthy subjects group was statistically significant. In the present article, we focused our attention on the role of single nucleotide polymorphisms (SNPs) of the components of the circadian system on melanoma biology. In particular, we intended to test the hypothesis that specific SNPs of the circadian clock genes, such as AANAT (arylalkylamine N-acetyltransferase), CLOCK (clock circadian regulator), NPAS2 (neuronal PAS domain protein 2), PER1 (period circadian clock 1), PER2 (period circadian clock 2), RORA (retinoic acid-related orphan receptor A), and TIMELESS (timeless circadian clock) could significantly influence the predisposition to develop cutaneous melanoma or the outcome of melanoma bearing patients. To this aim, we genotyped 629 cases of melanoma and 610 healthy controls in 14 known SNPs of the above reported clock genes.
Material and methods
Study design
We conducted a retrospective study to test the hypothesis that genetic variants, such as SNPs, of the circadian pathway might be associated with the susceptibility or prognosis of patients affected with melanoma. To this aim, we extracted the clinico-pathological data of patients treated at our institution (Veneto Institute of Oncology, Italy) between 2008 and 2015, using a prospectively maintained database linked to our institutional biobank (Clinica Chirurgica I—University Hospital of Padova, Italy). To be included in the study, each case had to meet the following requirements: (1) histologically confirmed diagnosis of cutaneous melanoma or metastasis from melanoma; (2) pathology-based information on TNM stage; (3) follow-up data (minimum follow up: six months); (4) availability of peripheral blood for genotyping purposes.
Patients and healthy donors
We retrospectively selected 629 consecutive melanoma patients, and 610 healthy controls. The selection of the latter was both population-based (n = 270 blood donors) and hospital-based (n = 340, healthy subjects who visited the Clinica Chirurgica I practice for routine check-ups). All patients signed an informed consent form explaining the research purposes of the blood withdrawal. Healthy controls dataset was already employed in our previous analyses [13, 14].
SNPs selection
We focused on 5 core clock genes, which are CLOCK, NPAS2, PER1, PER2, and RORA. Moreover, we added two clock-related genes TIMELESS, associated with cancer risk in several studies [15,16,17,18] and AANAT for its role in the biosynthesis of melatonin, a key marker of the circadian system.
We selected either Tag SNPs, interrogating The Genome Variation Server of the University of Washington (http://gvs.gs.washington.edu/GVS/) and the TagSNP tool of the US National Institute of Environmental Health Sciences (https://snpinfo.niehs.nih.gov/snpinfo/snptag.html), or variants already known to be associated with cancer susceptibility or prognosis, that had a minor allele frequency > 5%. We relied on our previous meta-analysis [19], on our previous case–control studies [13, 14, 20] and on literature. The publications referring to cancer studies for each SNP are listed in Table 2.
DNA extraction and genotyping as described in [13].
Statistical analysis
For susceptibility of melanoma assessment, multivariate logistic regression was employed, modeled additively by minor allele count. Odds ratios (OR) and 95% confidence intervals were used as a measure of association of each SNP. In those multivariate models, the evaluated outcome was the presence or absence of melanoma, while the explanatory variables were the single SNPs adjusted for age and gender.
Subgroup analysis was performed by gender. For the female subgroup, a further discrimination was performed by age (> 50 years old vs < 50 years old).
Sensitivity analysis was conducted excluding patients with melanoma of unknown primary (MUP).
For prognosis of melanoma assessment, multivariate Cox proportional hazard regression was employed. Overall survival was defined as the time from the date of tumor diagnosis to the date of death by any cause or last follow-up visit. Hazard ratios (HR) and 95% confidence intervals were used as a measure of association. In those multivariate models the evaluated event was the patient’s survival, the time to event were the months of survival, and the explanatory variables were the single SNP adjusted for age, gender and melanoma stage.
The Benjamini–Hochberg method (1995) was employed as adjustment for multiple comparisons (False Discovery Rate Online Calculator, 2016, Carbocation Corporation, https://tools.carbocation.com/FDR). False discovery rate (FDR) cut-off was set at 0.1.
Hardy-Weimberg Equilibrium (HWE) was tested for both samples (patients and healthy controls) for each SNP employing OEGE—Online Encyclopedia for Genetic Epidemiology studies [26], http://www.oege.org/software/hwe-mr-calc.shtml. This tool is a HWE calculator for biallelic SNPs based on Chi—square statistic.
Statistical power was calculated for each SNP employing the on-line tool “Power and Sample Size” of the University of Vanderbilt (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize) [27]. Power was defined as the probability of correctly rejecting the null hypothesis that the relative risk (OR) was equal to 1, given 629 case patients and 610 controls. The type I error probability α was set to 0.05 and ψ (OR considered clinically relevant in our study) was set to 0.80. Finally, the expression quantitative trait locus (eQTL) analysis was carried out interrogating the GTEx Portal (https://gtexportal.org/home/).
Rcmdr: R Commander. R package version 2.4–2 was employed for the analyses.
Results
The analysis was based on a total of 1239 subjects, 629 cases of melanoma and 610 healthy controls, all of European ancestry. Thirty-four out of 629 patients had melanoma of unknown primary. The healthy control dataset was already employed in our previous analyses [13, 14], detailed subjects’ characteristics and the main features of the SNPs we investigated are reported in Tables 1 and 2, respectively.
A total of 14 preselected SNPs in 7 circadian clock genes were successfully genotyped, and no departures from Hardy–Weinberg equilibrium were observed neither among the controls nor among the patients, as summarized in Additional file 1: Table S1. The mean statistical power for this analysis was approximately 70%; detailed statistical power for each SNP is reported in Additional file 1: Table S1.
Susceptibility and prognosis assessment
Associations between the selected clock gene SNPs and melanoma susceptibility were tested assuming an additive model of inheritance. We used odds ratios (ORs) and their corresponding 95% confidence intervals (95% CI) to measure the strength of association between each polymorphism and melanoma susceptibility. The results are reported in Table 3.
After adjusting for multiple testing, we identified two SNPs on RORA locus significantly associated with melanoma susceptibility. rs339972 C allele and rs10519097 T allele were associated with a decreased melanoma risk (OR 0.76; 95% CI 0.63–0.91; P = 0.003 and OR 0.77; 95% CI 0.61–0.97; P = 0.026, respectively). Adjusted p-values are reported in Additional file 1: Table S2.
No differences were found excluding from the analyses the 34 patients with melanoma of unknown primary.
Considering the putative role of RORA as a nuclear steroid hormone receptor, we decided to conduct a subgroup analysis by gender. The results are reported in Table 4.
Interestingly, the RORA rs339972 C allele was associated with a decreased melanoma susceptibility only in the female subgroup (OR 0.67; 95% CI 0.51–0.88; P = 0.003) while RORA rs10519097 T allele was associated with a decreased predisposition to develop melanoma only in the male subgroup (OR 0.62; 95% CI 0.44–0.87; P = 0.005). A further subgroup analysis was carried out dividing the female dataset by age at diagnosis (> 50 years old vs < 50 years old) to discriminate if menopausal or premenopausal status could interact with RORA variants and melanoma associations. Females aged over 50 years old carrying RORA rs339972 C allele had a decreased susceptibility to develop melanoma of 33% (OR 0.67; 95% CI 0.54–0.83; P = 0.0002). No other tested associations were statistically significant after correcting for multiple testing.
No statistically significant associations with prognosis were observed as reported, for each SNP, in Additional file 1: Table S3.
eQTL search
The expression quantitative trait locus (eQTL) analysis was carried out interrogating the GTEx Portal. A significant result (P = 0.000015) was found for RORA rs339972 in pancreas, while no significant eQTLs were identified for RORA SNP rs10519097.
Discussion
Summary
In the present study, we tested the hypothesis that DNA genetic variations of the circadian clock genes might influence the susceptibility to develop cutaneous melanoma or the outcome of melanoma patients. The hypothesis stemmed from the growing number of studies correlating the risk or the prognosis of different tumour types with the genetic variability of the circadian system [19]. Moreover, cutaneous melanoma and the circadian system share a common denominator: ultraviolet (UV) radiation. The persistent exposure to sunlight, resulting in sunburn, especially in childhood, is an acknowledged melanoma risk factor [45]. Of note, light is the principal zeitgeber (literally time giver) of the biological clock, and thus plays a key role in clock synchronization. We selected 14 single nuclear polymorphisms in seven clock-related genes and we genotyped 1239 subjects, 629 melanoma patients and 610 healthy donors. Our results indicated that two SNPs of the RORA clock gene are associated with melanoma susceptibility. In particular, subgroup analysis revealed that rs339972 was statistically significant with respect to the association with melanoma susceptibility in females, while rs10519097 in males. Moreover, females carrying RORA rs339972 C allele aged over 50 years had a decreased susceptibility to develop melanoma of 33%. No associations were found with melanoma prognosis.
RORA structure
RORA (also known as NR1F1, nuclear receptor subfamily 1, group F, member 1) belongs to the nuclear steroid hormone receptor family of transcription factors. It was identified in the 1990s for its sequence similarity with the retinoid acid receptor (RAR) [46, 47]. Receptors without their ligands were called orphan-receptors. More recently, the crystal structure of its ligand binding domain identified cholesterol and cholesterol sulphate as putative ligands [48, 49]. The RORA locus maps to human chromosome 15q22.2 and spans 730 kb genomic region, comprised of 15 exons. Alternative splicing and alternative promoter usage generate four isoforms RORA1-4, differently expressed in a tissue-specific manner. Different isoforms of RORA have different binding specificities and strengths of transcriptional activity [47]. In the early 2000s RORA was shown to display a rhythmic pattern of expression in a circadian manner in the liver, kidney, retina, lung and suprachiasmatic nuclei, the region of the brain responsible for circadian rhythmicity control. Few years later, RORA was shown to be necessary for the rhythmic circadian behavior, indeed mice deficient in RORA display an aberrant circadian rhythmicity [50]. In the clock machinery, RORA competes with other nuclear receptors REV-ERBα and β (NR1D1 and 2, nuclear receptor subfamily 1, group D, member 1 and 2, respectively) to bind specific DNA response element (RORE) in the promoter of the core clock gene BMAL1 (brain and muscle Arnt-like protein-1, also known as ARNTL1); RORA activates, while REV-ERBα and β suppress BMAL1 transcription [51,52,53]. Moreover, melatonin, a major clock output, putatively regulates RORA [54].
Both RORA studied variants, rs10519097 and rs33997, are located within introns (first and second respectively) of RORA transcript 1, referred to as RORA1, while they are upstream to transcripts 2–4, RORA2-4.
Circadian clock and melanoma
Approximately ten years ago, a circadian clock was found also within the skin [55], in particular the different cellular subtypes, fibroblasts, keratinocyte and melanocytes have a local circadian machinery which includes the expression of RORA [56]. In cutaneous melanoma it has been described a general reduction of clock genes abundance compared to normal adjacent tissues [8]. RORA was found to be downregulated in melanoma compared to nevi, and the expression was directly correlated to overall and disease-free survival [7]. Moreover, non-metastatic cutaneous melanoma induces chronodisruption in central and peripheral circadian clocks [9]. Recently, two variants in the RORA locus were found to be associated with melanoma prognosis, rs782917 and rs17204952 as well as rs7253062 in DNMT1, a steroid hormone receptor as well. Combined analysis of risk genotypes of the three variants revealed a decreased cutaneous melanoma specific survival in a dose–response manner. The authors performed a pathway-based analysis to evaluate genetic variants of 191 steroid hormone-related genes [11]. This research rationale was based on the shared idea that melanoma represent a steroid-hormone related malignancy [57].
Sex hormones and melanoma
Beside the mentioned risk factors, additional variables are ethnicity, age and gender, in the latter a female advantage has been generally revealed. Lifestyle, X chromosome dosage and sex hormones play a role in this disparity [58]. It has been reported that incidence rates of cutaneous melanoma rise steeply in women until about age 50, suggesting estrogen as a possible risk factor. Moreover, a cumulative dose-dependent increased risk of cutaneous melanoma was shown with the use of estrogens, oral contraceptives and hormonal replacement therapy, [59] and the risk of premenopausal melanoma may be increased among women who are current oral contraceptive users, particularly among those with longer durations of use [60]. An epidemiological study reported that the incidence of pregnancy-associated melanoma increases with increasing maternal age (women aged 40–55 had a 7.55-fold higher risk than women aged 15–24) [61]. Women with a severe teenage acne history, reflecting a hormone imbalance, as a higher circulating level of free testosterone, had an increased relative risk of melanoma [62]. In males, personal history of prostate cancer, neoplasia in which androgens play a major role, was found to be associated with an increased risk of melanoma [63].
Sexually dimorphic regulation of RORA
Male and female sex hormones differentially and reciprocally regulate RORA, with androgen suppressing RORA while estrogen enhancing RORA expression. Dihydrotestosterone (DHT) and estradiol increase the binding of androgen receptor (AR) and estrogen receptor (ER) respectively to the RORA promoter region, and RORA in turn increases testosterone levels. The resultant negative feedback transcriptionally regulates aromatase, which converts male hormones to estrogens [64].
Possible RORA SNPs interactions with melanoma susceptibly in a gender-specific manner
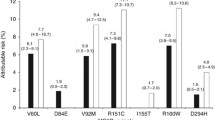
The results of the present study are supporting the role of hormone receptors in melanoma susceptibility. In particular, the analyzed RORA polymorphisms have different effect on melanoma susceptibility whereas rs339972 minor allele has a protective effect on premenopausal women while rs10519097 minor allele has a protective effect exclusively in men. A speculation could be that those polymorphisms interact differently with sex hormones and RORA expression, given the general knowledge that clock genes as well as RORA are down regulated in cancer including melanoma. Another hypothesis might be that the effect of different genetic variations is related to different RORA isoforms or to different RORA functions; beside the involvement in circadian rhythmicity, RORs molecules play a critical role in development, immunity and cellular metabolism [53]. Further information on these genetic variants derives from our previous studies. In our previous meta-analysis [19], rs339972 was statistically significant with respect to the association with cancer in general while rs10519097 with breast cancer risk. In our previous case–control studies [13, 14, 20] we found rs339972 to be associated with both gastric cancer and sarcoma susceptibility. In all those studies, the minor allele C had a protective effect, but the associations were statistically less robust (in terms of P-value) as compared to the association revealed by the present work. Noticeably, in our previous meta-analysis subgroups meta-analysis employing two datasets (4587 subjects) showed that the association of rs10519097 with breast cancer was significant with an intermediate level of evidence (summary OR: 0.85, CI: 0.75–0.96, P = 0.008). The A allele was associated with a reduced risk of developing cancer also in postmenopausal breast cancer patients (3700 subjects; summary OR: 0.87, CI: 0.75–1, P = 0.04) [19]. Nonetheless, further studied will be needed to explain and understand the mechanisms underlying our results.
The GTEx Portal was queried for the expression quantitative trait locus (eQTL) analysis. A significant result (P = 0.000015) was found for RORA rs339972 in pancreas, nevertheless a GWAS [65] revealed no significant associations with pancreatic cancer risk (OR: 0.92, CI: 0.85–1.00, P = 0.04). Further analyses are necessary to unravel the biological and clinical significance of those results.
Conclusions
To the best of our knowledge, this is one of the few pioneering analyses investigating the relationship between cutaneous melanoma susceptibility or prognosis and circadian gene variants. Nonetheless, we must acknowledge some limitations of our work. First, we tested a limited number of clock genes and for each of them evaluated only selected SNPs, based on our previous analyses or on literature, therefore studies involving a larger amount of genes and SNPs are eagerly awaited. Second, all the enrolled subjects were of Caucasian ethnicity. Additional studies including multiple ethnicity should be performed in order to validate our hypothesis also in people with different ancestry. Third, the source of controls was both population and hospital based (University Hospital of Padova, Italy), nevertheless we choose patients with different conditions for the hospital-based fraction (i.e. patients treated for haemorrhoids, goitre, gastritis) to avoid selection bias. Fourth, we employed only one genetic model (that is, the additive model), whereas neither the recessive nor the dominant model were explored: however, our aim was not to identify the best genetic model for specific polymorphisms but rather to summarize (in a quantitative fashion) the evidence regarding the selected genetic variants.
Overall, we cannot conclude in favour of the hypothesis that variability of the circadian clock genes may affect melanoma susceptibility or prognosis, but we did find an interesting relationship between melanoma biology and RORA gene variants, which we believe is worth further investigation.
Availability of data and materials
All data generated or analysed during this study are included in this published article [and its additional information files].
Abbreviations
- SNP:
-
Single nucleotide polymorphism
- GWAS:
-
Genome-wide association studies
- NGS:
-
Next generation sequencing
- IARC:
-
International Agency for Research on Cancer
- OR:
-
Odds ratios
- HR:
-
Hazard ratios: 95%CI: 95% confidence interval
- MUP:
-
Melanoma of unknown primary
- eQTL:
-
Expression quantitative trait locus
- FDR:
-
False discovery rate
- HWE:
-
Hardy-Weimberg Equilibrium
- Chr:
-
Chromosome
- RAR:
-
Retinoid acid receptor
- DHT:
-
Dihydrotestosterone
- AR:
-
Androgen receptor
- ER:
-
Estrogen receptor
References
Papakostas D, Stefanaki I, Stratigos A. Genetic epidemiology of malignant melanoma susceptibility. Melanoma Manag. 2015;2:165–9.
Rossi M, Pellegrini C, Cardelli L, Ciciarelli V, Di Nardo L, Fargnoli MC. Familial melanoma: diagnostic and management implications. Dermatol Pract Concept. 2019;9:10–6.
Pasquali S, Hadjinicolaou AV, Chiarion Sileni V, Rossi CR, Mocellin S. Systemic treatments for metastatic cutaneous melanoma. Cochrane Database Syst Rev. 2018;2:CD011123.
Buja A, Lange JH, Perissinotto E, Rausa G, Grigoletto F, Canova C, et al. Cancer incidence among male military and civil pilots and flight attendants: an analysis on published data. Toxicol Ind Health. 2005;21:273–82.
Buja A, Mastrangelo G, Perissinotto E, Grigoletto F, Frigo AC, Rausa G, et al. Cancer incidence among female flight attendants: a meta-analysis of published data. J Womens Health (Larchmt). 2006;15:98–105.
Straif K, Baan R, Grosse Y, Secretan B, El Ghissassi F, Bouvard V, et al. Carcinogenicity of shift-work, painting, and fire-fighting. Lancet Oncol. 2007;8:1065–6.
Brożyna AA, Jóźwicki W, Skobowiat C, Jetten A, Slominski AT. RORα and RORγ expression inversely correlates with human melanoma progression. Oncotarget. 2016;7:63261–82.
Lengyel Z, Lovig C, Kommedal S, Keszthelyi R, Szekeres G, Battyani Z, et al. Altered expression patterns of clock gene mRNAs and clock proteins in human skin tumors. Tumour Biol. 2013;34:811–9.
de Assis LVM, Moraes MN, Magalhães-Marques KK, Kinker GS, da Silveira C-M, Castrucci AML. Non-metastatic cutaneous melanoma induces chronodisruption in central and peripheral circadian clocks. Int J Mol Sci. 2018;19:1065. https://doi.org/10.3390/ijms19041065.
Franzoni A, Markova-Car E, Dević-Pavlić S, Jurišić D, Puppin C, Mio C, et al. A polymorphic GGC repeat in the NPAS2 gene and its association with melanoma. Exp Biol Med (Maywood). 2017;242:1553–8.
Li B, Wang Y, Xu Y, Liu H, Bloomer W, Zhu D, et al. Genetic variants in RORA and DNMT1 associated with cutaneous melanoma survival. Int J Cancer. 2018;142:2303–12.
Rocca MS, Benna C, Mocellin S, Rossi CR, Msaki A, Di Nisio A, et al. E2F1 germline copy number variations and melanoma susceptibility. J Transl Med. 2019;17:181.
Benna C, Rajendran S, Spiro G, Tropea S, Del Fiore P, Rossi CR, et al. Associations of clock genes polymorphisms with soft tissue sarcoma susceptibility and prognosis. J Transl Med. 2018;16:338.
Rajendran S, Benna C, Marchet A, Nitti D, Mocellin S. Germline polymorphisms of circadian genes and gastric cancer predisposition. Cancer Commun (Lond). 2020.
Fu A, Leaderer D, Zheng T, Hoffman AE, Stevens RG, Zhu Y. Genetic and epigenetic associations of circadian gene TIMELESS and breast cancer risk. Mol Carcinog. 2012;51:923–9.
Grundy A, Schuetz JM, Lai AS, Janoo-Gilani R, Leach S, Burstyn I, et al. Shift work, circadian gene variants and risk of breast cancer. Cancer Epidemiol. 2013;37:606–12.
Jim HS, Lin HY, Tyrer JP, Lawrenson K, Dennis J, Chornokur G, et al. Common genetic variation in circadian rhythm genes and risk of epithelial ovarian cancer (EOC). J Genet Genome Res. 2015;2:017 (Epub 2015 Sep 15).
Zhu Y, Stevens RG, Hoffman AE, Fitzgerald LM, Kwon EM, Ostrander EA, et al. Testing the circadian gene hypothesis in prostate cancer: a population-based case-control study. Cancer Res. 2009;69:9315–22.
Benna C, Helfrich-Forster C, Rajendran S, Monticelli H, Pilati P, Nitti D, et al. Genetic variation of clock genes and cancer risk: a field synopsis and meta-analysis. Oncotarget. 2017;8:23978–95.
Rajendran S, Benna C, Monticelli H, Spiro G, Menin C, Mocellin S. Germline variation of circadian pathway genes and prognosis of gastric cancer patients. Gut. 2018;67:779–80.
Dai H, Zhang L, Cao M, Song F, Zheng H, Zhu X, et al. The role of polymorphisms in circadian pathway genes in breast tumorigenesis. Breast Cancer Res Treat. 2011;127:531–40.
Karantanos T, Theodoropoulos G, Gazouli M, Vaiopoulou A, Karantanou C, Stravopodis DJ, et al. Association of the clock genes polymorphisms with colorectal cancer susceptibility. J Surg Oncol. 2013;108:563–7.
Li J, Humphreys K, Heikkinen T, Aittomaki K, Blomqvist C, Pharoah PD, et al. A combined analysis of genome-wide association studies in breast cancer. Breast Cancer Res Treat. 2011;126:717–27.
Truong T, Liquet B, Menegaux F, Plancoulaine S, Laurent-Puig P, Mulot C, et al. Breast cancer risk, nightwork, and circadian clock gene polymorphisms. Endocr Relat Cancer. 2014;21:629–38.
Zhao B, Lu J, Yin J, Liu H, Guo X, Yang Y, et al. A functional polymorphism in PER3 gene is associated with prognosis in hepatocellular carcinoma. Liver Int. 2012;32:1451–9.
Rodriguez S, Gaunt TR, Day IN. Hardy-Weinberg equilibrium testing of biological ascertainment for Mendelian randomization studies. Am J Epidemiol. 2009;169:505–14.
Dupont WD, Plummer WD Jr. Power and sample size calculations. A review and computer program. Control Clin Trials. 1990;11:116–28.
Zienolddiny S, Haugen A, Lie JA, Kjuus H, Anmarkrud KH, Kjaerheim K. Analysis of polymorphisms in the circadian-related genes and breast cancer risk in Norwegian nurses working night shifts. Breast Cancer Res. 2013;15:R53.
Monsees GM, Kraft P, Hankinson SE, Hunter DJ, Schernhammer ES. Circadian genes and breast cancer susceptibility in rotating shift workers. Int J Cancer. 2012;131:2547–52.
Garufi,Carlo, Giacomini,Elisa, Torsello,Angela, Sperduti,Isabella, Melucci,Elisa, Mottolese,Marcella, et al. Gender effects of single nucleotide polymorphisms and miRNAs targeting clock-genes in metastatic colorectal cancer patients (mCRC).
Hoffman AE, Yi CH, Zheng T, Stevens RG, Leaderer D, Zhang Y, et al. CLOCK in breast tumorigenesis: genetic, epigenetic, and transcriptional profiling analyses. Cancer Res. 2010;70:1459–68.
Miki I, Tamura T, Nakamura T, Makimoto H, Hamana N, Uchiyama H, et al. Circadian variability of pharmacokinetics of 5-fluorouracil and CLOCK T3111C genetic polymorphism in patients with esophageal carcinoma. Ther Drug Monit. 2005;27:369–74.
Okuno T, Tamura T, Yamamori M, Chayahara N, Yamada T, Miki I, et al. Favorable genetic polymorphisms predictive of clinical outcome of chemoradiotherapy for stage II/III esophageal squamous cell carcinoma in Japanese. Am J Clin Oncol. 2007;30:252–7.
Zhou F, He X, Liu H, Zhu Y, Jin T, Chen C, et al. Functional polymorphisms of circadian positive feedback regulation genes and clinical outcome of Chinese patients with resected colorectal cancer. Cancer. 2012;118:937–46.
Wang B, Dai ZM, Zhao Y, Wang XJ, Kang HF, Ma XB, et al. Current evidence on the relationship between two common polymorphisms in NPAS2 gene and cancer risk. Int J Clin Exp Med. 2015;8:7176–83.
Zhu Y, Stevens RG, Leaderer D, Hoffman A, Holford T, Zhang Y, et al. Non-synonymous polymorphisms in the circadian gene NPAS2 and breast cancer risk. Breast Cancer Res Treat. 2008;107:421–5.
Zhu Y, Leaderer D, Guss C, Brown HN, Zhang Y, Boyle P, et al. Ala394Thr polymorphism in the clock gene NPAS2: a circadian modifier for the risk of non-Hodgkin’s lymphoma. Int J Cancer. 2007;120:432–5.
Chu L, Zhu Y, Yu K, Zheng T, Yu H, Zhang Y, et al. Variants in circadian genes and prostate cancer risk: a population-based study in China. Prostate Cancer Prostatic Dis. 2008;11:342–8.
Markt SC, Valdimarsdottir UA, Shui IM, Sigurdardottir LG, Rider JR, Tamimi RM, et al. Circadian clock genes and risk of fatal prostate cancer. Cancer Causes Control. 2015;26:25–33.
Yi C, Mu L, de la Longrais IA, Sochirca O, Arisio R, Yu H, et al. The circadian gene NPAS2 is a novel prognostic biomarker for breast cancer. Breast Cancer Res Treat. 2010;120:663–9.
Yuan P, Wang S, Zhou F, Wan S, Yang Y, Huang X, et al. Functional polymorphisms in the NPAS2 gene are associated with overall survival in transcatheter arterial chemoembolization-treated hepatocellular carcinoma patients. Cancer Sci. 2014;105:825–32.
Rana S, Shahid A, Ullah H, Mahmood S. Lack of association of the NPAS2 gene Ala394Thr polymorphism (rs2305160:G>A) with risk of chronic lymphocytic leukemia. Asian Pac J Cancer Prev. 2014;15:7169–74.
Madden MH, Anic GM, Thompson RC, Nabors LB, Olson JJ, Browning JE, et al. Circadian pathway genes in relation to glioma risk and outcome. Cancer Causes Control. 2014;25:25–32.
Zhang Z, Ma F, Zhou F, Chen Y, Wang X, Zhang H, et al. Functional polymorphisms of circadian negative feedback regulation genes are associated with clinical outcome in hepatocellular carcinoma patients receiving radical resection. Med Oncol. 2014;31:179 (Epub 2014 Oct 26).
Leiter U, Keim U, Garbe C. Epidemiology of Skin Cancer: Update 2019. Adv Exp Med Biol. 2020;1268:123–39.
Becker-André M, André E, DeLamarter JF. Identification of nuclear receptor mRNAs by RT-PCR amplification of conserved zinc-finger motif sequences. Biochem Biophys Res Commun. 1993;194:1371–9.
Giguère V, Tini M, Flock G, Ong E, Evans RM, Otulakowski G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8:538–53.
Kallen J, Schlaeppi JM, Bitsch F, Delhon I, Fournier B. Crystal structure of the human RORalpha Ligand binding domain in complex with cholesterol sulfate at 2.2 A. J Biol Chem. 2004;279:14033–8.
Kallen JA, Schlaeppi JM, Bitsch F, Geisse S, Geiser M, Delhon I, et al. X-ray structure of the hRORalpha LBD at 1.63 A: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORalpha. Structure. 2002;10:1697–707.
Akashi M, Takumi T. The orphan nuclear receptor RORalpha regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12:441–8.
Burris TP. Nuclear hormone receptors for heme: REV-ERBalpha and REV-ERBbeta are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22:1509–20.
Ueda HR, Chen W, Adachi A, Wakamatsu H, Hayashi S, Takasugi T, et al. A transcription factor response element for gene expression during circadian night. Nature. 2002;418:534–9.
Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003.
Dai J, Ram PT, Yuan L, Spriggs LL, Hill SM. Transcriptional repression of RORalpha activity in human breast cancer cells by melatonin. Mol Cell Endocrinol. 2001;176:111–20.
Tanioka M, Yamada H, Doi M, Bando H, Yamaguchi Y, Nishigori C, et al. Molecular clocks in mouse skin. J Invest Dermatol. 2009;129:1225–31.
de Assis LVM, Moraes MN, Castrucci AML. The molecular clock in the skin, its functionality, and how it is disrupted in cutaneous melanoma: a new pharmacological target? Cell Mol Life Sci. 2019;76:3801–26.
Mitkov M, Joseph R, Copland J. Steroid hormone influence on melanomagenesis. Mol Cell Endocrinol. 2015;417:94–102.
Bellenghi M, Puglisi R, Pontecorvi G, De Feo A, Carè A, Mattia G. Sex and Gender Disparities in Melanoma. Cancers (Basel). 2020;12:1819. https://doi.org/10.3390/cancers12071819.
Koomen ER, Joosse A, Herings RM, Casparie MK, Guchelaar HJ, Nijsten T. Estrogens, oral contraceptives and hormonal replacement therapy increase the incidence of cutaneous melanoma: a population-based case-control study. Ann Oncol. 2009;20:358–64.
Feskanich D, Hunter DJ, Willett WC, Spiegelman D, Stampfer MJ, Speizer FE, et al. Oral contraceptive use and risk of melanoma in premenopausal women. Br J Cancer. 1999;81:918–23.
Bannister-Tyrrell M, Roberts CL, Hasovits C, Nippita T, Ford JB. Incidence and outcomes of pregnancy-associated melanoma in New South Wales 1994–2008. Aust N Z J Obstet Gynaecol. 2015;55:116–22.
Zhang M, Qureshi AA, Fortner RT, Hankinson SE, Wei Q, Wang LE, et al. Teenage acne and cancer risk in US women: A prospective cohort study. Cancer. 2015;121:1681–7.
Li WQ, Qureshi AA, Ma J, Goldstein AM, Giovannucci EL, Stampfer MJ, et al. Personal history of prostate cancer and increased risk of incident melanoma in the United States. J Clin Oncol. 2013;31:4394–9.
Sarachana T, Xu M, Wu RC, Hu VW. Sex hormones in autism: androgens and estrogens differentially and reciprocally regulate RORA, a novel candidate gene for autism. PLoS ONE. 2011;6:e17116.
Petersen GM, Amundadottir L, Fuchs CS, Kraft P, Stolzenberg-Solomon RZ, Jacobs KB, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42:224–8.
Acknowledgements
We acknowledge: The personnel of the Biobank of the 1st Surgical Clinic, University Hospital of Padova—Istituto Oncologico Veneto (IOV-IRCCS) Padova, Italy. The personnel of the Istituto Oncologico Veneto (IOV-IRCCS, Padova, Italy) Manuela Ruffato and Sonia Nalesso for organizing the sampling activity and the informed consent retrieval; Paolo Del Fiore and Romina Spina for managing clinico-pathological data and the Ethic Committee requests. “Piccoli Punti”, an Italian no-profit association for melanoma research and prevention, for entirely sustaining GS grant and part of the research. UNICREDIT BANCA for donating laboratory equipment useful for the present research. Dr. MS Rocca, Department of Medicine, Unit of Andrology and Reproductive Medicine, University of Padua, Padua, Italy, for the fruitful suggestions regarding the manuscript.
Funding
University of Padova, BIRD168075, “Germline polymorphisms of candidate genes as predictor of risk and prognosis in patients with cutaneous melanoma and soft tissue sarcoma”. Istituto Oncologico Veneto (IOV-IRCCS), 5 × 1000, BIGID219MOCE.
Author information
Authors and Affiliations
Contributions
CB: Conceptualization, Formal analysis, Writing—Original Draft; SR, GS: Investigation, Validation; LDO: Resources; CM: Writing—Review & Editing; SM, CRR: Supervision; Funding acquisition. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Padova University Hospital Ethic Committee prot#448.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Hardy-Weinberg equilibrium (HWE) test and detailed statistical power for each SNP analysis. Table S2. Benjamini-Hochberg adjusted p-values for each SNP analysis. Table S3. Associations of circadian pathway genes with prognosis of 629 melanoma patients under the additive model of inheritance.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Benna, C., Rajendran, S., Spiro, G. et al. Gender-specific associations between polymorphisms of the circadian gene RORA and cutaneous melanoma susceptibility. J Transl Med 19, 57 (2021). https://doi.org/10.1186/s12967-021-02725-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-021-02725-5