Abstract
Background
Kallmann syndrome (KS) is a common type of idiopathic hypogonadotropic hypogonadism. To date, more than 30 genes including ANOS1 and FGFR1 have been identified in different genetic models of KS without affirmatory genotype–phenotype correlation, and novel mutations have been found.
Methods
A total of 35 unrelated patients with clinical features of disorder of sex development were recruited. Custom-panel sequencing or whole-exome sequencing was performed to detect the pathogenic mutations. Sanger sequencing was performed to verify single-nucleotide variants. Copy number variation-sequencing (CNV-seq) was performed to determine CNVs. The pathogenicity of the identified variant was predicted in silico. mRNA transcript analysis and minigene reporter assay were performed to test the effect of the mutation on splicing.
Results
ANOS1 gene c.709 T > A and c.711 G > T were evaluated as pathogenic by several commonly used software, and c.1063-2 A > T was verified by transcriptional splicing assay. The c.1063-2 A > T mutation activated a cryptic splice acceptor site downstream of the original splice acceptor site and resulted in an aberrant splicing of the 24-basepair at the 5′ end of exon 8, yielding a new transcript with c.1063–1086 deletion. FRFR1 gene c.1835delA was assessed as pathogenic according to the ACMG guideline. The CNV of del(8)(p12p11.22)chr8:g.36140000_38460000del was judged as pathogenic according to the ACMG & ClinGen technical standards.
Conclusions
Herein, we identified three novel ANOS1 mutations and two novel FGFR1 variations in Chinese KS families. In silico prediction and functional experiment evaluated the pathogenesis of ANOS1 mutations. FRFR1 c.1835delA mutation and del(8)(p12p11.22)chr8:g.36140000_38460000del were assessed as pathogenic variations. Therefore, our study expands the spectrum of mutations associated with KS and provides diagnostic evidence for patients who carry the same mutation in the future.
Similar content being viewed by others
Background
Idiopathic hypogonadotropic hypogonadism (IHH) is a rare genetic disease characterized by delayed or complete lack of puberty, which is caused by hypothalamic-pituitary–gonadal (HPG) axis dysfunction [1]. IHH is clinically divided into Kallmann syndrome with anosmia/hyposmia (KS) and normosmic IHH (nIHH). KS accounts for 50% of all IHH cases [2], and KS occurs in 1 per 30,000 men and 1 per 125,000 women [3].
KS is clinically heterogeneous, mainly manifesting as gonadal dysplasia and possibly accompanied by other congenital malformations, such as renal agenesis or hypoplasia, cleft lip/palate, dental agenesis, hearing impairment, bimanual synkinesis, or skeletal anomalies [1]. KS exhibits different inheritance patterns and genetic heterogeneity including X-linked recessive (ANOS1), autosomal recessive (PROK2 and PROKR2), and autosomal dominant (FGFR1, FGF8, and CHD7) genes [4, 5]. ANOS1, located at the Xp22.3 and also known as Kallmann syndrome 1 (KAL1), is the first gene found to be associated with the X-linked form of KS [6]. Anosmin-1 protein that is encoded by ANOS1 contains four domains: a N-terminal cysteine-rich region (CR domain), a whey acidic protein-like (WAP) domain, four fibronectin type III (FnIII) domains, and a histidine-rich C-terminal region [7]. Currently, more than 200 mutations in ANOS1 have been included in the Human Gene Mutation Database (HGMD), all of which spread on the whole gene, and the number of mutation is increasing, but no mutation hotspots have been found in the affected regions [8].
Fibroblast growth factor receptor 1 (FGFR1) is located at 8p11.23. FGFR1 is a member of the tyrosine kinase superfamily of receptors. Dodé et al. reported that loss-of-function mutations of FGFR1 underlay KS2 for the first time in 2003. KS2 is an autosomal dominant disease [9]. To date, more than 300 mutations in FGFR1 have been included in HGMD, all of which included missense mutation, nonsense mutation, splice mutation, and rarely deletions. FGFR1 signaling plays an important role in neuronal migration, differentiation, survival, as well as cell proliferation during embryonic development mainly via the PI3K/Akt and MAPK pathways [10].
In this study, we recruited 35 unrelated individuals with IHH manifestations and identified three novel mutations in ANOS1 and two novel mutations in FGFR1. Additionally, we verified the novel splice site mutation for transcription function; the c.1063-2 A > T mutation generated an abnormal transcript and translated into a truncated protein product. Thus far, more than 30 genes linked with IHH pathogenesis have been identified; however, the mutation spectrum is incomplete owing to the high genetic and clinical heterogeneity of KS [11]. Thus, our study expanded the mutation spectrum of KS and provided diagnostic evidence for patients with the same mutation in the future.
Methods
Participants
In total, 35 unrelated patients were recruited from Shengjing Hospital of China Medical University, all patients came for consultation because of disorder of sex development (DSD) (Table S1). In family 1, the proband (patient 1) manifested as small penis, cryptorchidism, and testicular dysplasia. In family 2, the proband (patient 2) phenotyped as small penis and testicular dysplasia. In family 3, the proband (patient 3) and his nephew showed cryptorchidism, left renal agenesis, and olfactory disorder. In family 4, the proband (patient 4) manifested as small penis, olfactory disorder, testicular dysplasia and polydactylism. In family 5, the proband (patient 5) phenotyped as cryptorchidism and testicular dysplasia.
Genetic testing
Genomic DNA was extracted from peripheral blood samples using a QIAamp DNA Blood Mini Kit (QIAGEN, Germany) according to the manufacturer’s instructions. Custom-panel sequencing (575 genes covering endocrine-related diseases) and whole-exome sequencing (WES) were performed by MyGenostics Inc. (Beijing, China) (Table S1). After sequencing, the raw data were saved as a FASTQ format. Both Illumina sequencing adapters and low quality reads (< 80 bp) were filtered by cutadaptor software (http://code.google.com/p/cutadapt/). The clean reads were mapped to the UCSC hg19 human reference genome using the parameter BWA of Sentieon software.(https://www.sentieon.com/). The duplicated reads were removed using the parameter driver of Sentieon software, and the parameter driver is used to correct the base, so that the quality value of the base in the reads of the final output BAM file can be closer to the real probability of mismatch with the reference genome, and the mapped reads were used for the detection of variation. The variants of SNP and InDel were detected by the parameter driver of Sentieon software. Then, the data would be transformed to VCF format. Variants were further annotated by ANNOVAR software (http://annovar.openbioinformatics.org/en/latest/). In the present study, three steps were used to select the potential pathogenic variants in downstream analysis: (1) variant reads should be more than 5, and variant ration should be no less than 30%; (2) the variants should be removed, when the highest minor allele frequency (MAF) in 1000 Genomes, EXAC, ESP6500, gnomAD, and dbSNP was more than 5%; (3) the synonymous mutations should be removed, when they were not in the HGMD database. After that, the rest mutations should be the potential pathogenic mutations for further analysis.
The candidate causative gene mutations were verified through the Sanger sequencing. Target gene fragments including mutations were amplified using the PrimeSTAR® HS DNA Polymerase (#R010Q, Takara, Dalian, China). Sanger sequencing was performed on 3730 DNA analyzer (Applied Biosystems, USA) using BigDye™ Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). Copy number variation-sequencing (CNV-seq) analysis was conducted using library construction kit (KR2000, Berry Genomics, Beijing, China) on the Illumina Nextseq CN500 platform. Sequencing data were analyzed by the data analysis system (Berry Genomics, Beijing, China).
ANOS1 mRNA transcript analysis
Total RNA was extracted from peripheral blood samples using a TransZol Up Plus RNA Kit (#ER501-01, TransGen Biotech Co., LTD, Dalian, Beijing, China) according to the manufacturer’s instructions. cDNA was synthesized from total RNA using PrimeScript™ RT reagent Kit with gDNA Eraser (#RR047A, Takara, Dalian, China). The cDNA products (NM_000216.4) were used as templates for PCR amplification using the following primers:
-
F: 5′—CGAGTGGCTGCTGTGAATGTG—3′
-
R: 5′—GCGAGTGGGTCGTCGTCTT—3′
ANOS1 mRNA transcripts were identified by Sanger sequencing.
Minigene reporter assay
The wild-type and variant amplicons encompassing intron 7, exon 8, and intron 8 of ANOS1 (NC_000023) were obtained from the genomic DNA of the father and proband, respectively, using the following primers:
-
F: 5′—CCGCTCGAGGCAGTCAGGAGCCACCGC—3′
-
R: 5′—CTAGCTAGCCTCTCCCTCCATTGTGCCTTG—3′
The PCR products were cloned into the pSPL3 vector, which was kindly provided by Professor Leping Shao, The Affiliated Qingdao Municipal Hospital of Qingdao University, China) by XhoI and NheI. Wild-type (pSPL3-ANOS1-WT) and variant (pSPL3-ANOS1-MUT) constructs were verified by Sanger sequencing (data not shown). Human embryo kidney cells (HEK293) were transfected with the plasmid of pSPL3-ANOS1-WT or pSPL3-ANOS1-MUT for 24 h using jetPEI®DNA transfection Reagent (Polyplus, France) according to the manufacturer’s protocol. cDNA was obtained from RNA extraction by reverse-transcription polymerase chain reaction (RT-PCR). The cDNA products were used as templates for PCR amplification using the following pSPL3 vector-specific primers [12]:
-
F: 5′—TCTGAGTCACCTGGACAACC—3′
-
R: 5′—ATCTCAGTGGTATTTGTGAGC—3′
Splicing transcripts were identified by Sanger sequencing.
Pathogenicity analysis of the variation
All mutations were assessed according to the American College of Medical Genetics and Genomics (ACMG) guideline. The pathogenicity of missense mutation was predicted using six tools: Revel (https://sites.google.com/site/revelgenomics), SIFT (http://sift.jcvi.org), PROVEAN (http://provean.jcvi.org), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2), MutationTaster (https://www.mutationtaster.org), M-CAP (http://bejerano.stanford.edu/mcap/), and GERP (http://mendel.stanford.edu/sidowlab/downloads/gerp/index.html). The evaluation of anosmin-1 amino acid conservation was performed by Ugene software [13] using the data from HomoloGene (ID:55,445). CNVs were evaluated according to the technical standards recommended by the ACMG and the Clinical Genome Resource (ClinGen).
Results
Monogene mutation screening and pedigree analysis
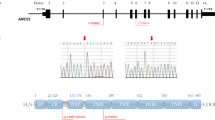
Five cases out of 35 unrelated patients were detected positive for mutations by custom-panel sequencing or WES, and their family members were analyzed. In family 1, a hemizygote ANOS1 (the transcript is NM_000216, hg19) mutation c.1063-2 A > T was found in the proband (Fig. 1A, II1), and the results of pedigree verification confirmed that the mutation was inherited from the proband’s mother (I2).
Pedigree and sequencing results of families 1–3. The pedigree showed core family members: square symbol represented males; circle symbol indicated females; Roman numerals represented generations; and Arabic numerals indicated the position of each individual within the family. A Pedigree and sequencing results of family 1. Sanger sequencing showed that the proband (II1) carried a hemizygous ANOS1 c.1063-2 A > T mutation, while the proband’s mother (I2) carried a heterozygous ANOS1 c.1063-2 A > T mutation. B Pedigree and sequencing results of family 2. NGS showed that the proband (II1) carried a hemizygous ANOS1 c.711 G > T mutation, while Sanger sequencing showed that the proband’s mother (I2) carried a heterozygous ANOS1 c.711 G > T mutation. C Pedigree and sequencing results of family 3. Sanger sequencing showed that the proband (II1) carried a hemizygous ANOS1 c.709 T > A mutation, the proband’s mother (I2) and sister (II2) carried a same heterozygous ANOS1 c.709 T > A mutation, and the proband’s nephew (III1) carried a hemizygote ANOS1 c.709 T > A mutation
In family 2, a hemizygote ANOS1 c.711 G > T mutation was found in the proband (Fig. 1B, II1), and the results of the pedigree verification confirmed that the mutation was inherited from the proband’s mother (I2).
In family 3, a hemizygote ANOS1 c.709 T > A mutation was found in the proband (Fig. 1C, II1). The results of the pedigree verification showed that the proband’s mother (I2) and sister (II2) carried heterozygous ANOS1 c.709 T > A mutation, and the proband’s nephew (III1) carried hemizygote ANOS1 c.709 T > A mutation.
In family 4, a de novo heterozygous FGFR1 (the transcript is NM_023110, hg19) mutation c.1835delA was found in the proband (Fig. 2A, II1), and the results of the pedigree verification showed that the father and mother were normal (I1 and I2).
Pedigree and sequencing results of families 4 and 5. A Pedigree and sequencing results of family 4. Sanger sequencing showed that the proband (II1) carried a heterozygous FGFR1 c.1835delA mutation, while the proband’s father (I1) and mother (I2) were normal. B Pedigree and CNV-seq results of family 5. CNV-seq results showed that the proband (II1) carried a 2.32 Mb deletion at 8p12-p11.22 (36,140,000–38,460,000), while the proband’s father (I1) and mother (I2) were normal
In family 5, the custom-panel sequencing result showed a sporadic 1.68 Mb deletion at 8p11.23-p11.22, and the CNV-seq results showed a 2.32 Mb deletion at 8p12-p11.22 (36,140,000–38,460,000) in the proband (Fig. 2B, II1), while the father and mother were normal (I1 and I2).
Verification of ANOS1 c.1063-2 A > T splicing mutation ex vivo and in vitro
To clarify the splicing effect of ANOS1 c.1063-2 A > T mutation in family 1, cDNA sequence analysis was performed ex vivo. As shown in Fig. 3, c.1063-2 A > T mutation resulted in an aberrant splicing of 24-bp at the 5′ end of exon 8 and generated a new transcript with c.1063–1086 deletion; both of the normal and aberrant transcripts of ANOS1 were detected in the proband’s peripheral blood (Fig. 3A). In addition, few aberrant transcripts were found in the maternal peripheral blood (Fig. 3A).
Results of the ANOS1 mRNA transcripts analysis. A Sanger sequencing results of the RT-PCR products showed existence of both the wild-type (WT) transcript and the mutant transcript in the peripheral blood. The mutant ANOS1 transcript was aberrant splicing with c.1063–1086 deletion. B Schematic structures of anosmin-1 proteins. The domains of anosmin-1 protein were retrieved from the Ensembl database (Transcript ID: ENST00000262648.8). The truncated protein exhibited a mutated FnIII.2 (ΔFnIII.2) domain with amino acid deletion at 355–362
Splicing experiment in vitro was performed by constructing a splicing reporter minigene vector. As shown in Fig. 4B, the RT-PCR product sequence of the mutation construct was shorter than that of the wild-type construct, and the sequencing analysis revealed that the c.1063-2 A > T mutation led to an aberrant splicing consistent with the validation ex vivo. As expected, a truncated protein with p.355–362 deletion was generated (Fig. 3B).
Results of minigene splicing assay. A Schematic diagram of minigene construction. The asterisk indicates the location of the c.1063-2 A > T mutation. The splicing pattern of wild-type (WT, top) and mutant (MUT, bottom) was showed respectively. B Minigene RT-PCR product sequencing results. The WT minigene formed normal mRNA composed of exon A, exon 8 and exon B. The mutant minigene caused a splicing abnormality, resulting in the 24 bp nucleotides deletion of 5′ end of exon 8 (c.1063–1086 deletion)
Pathogenicity analysis of ANOS1 c.709 T > A and c.711 G > T missense mutations
ANOS1 c.711 G > T and c.709 T > A variations in family 2 and 3 were two missense mutations that led to different amino acid substitutions of p.W237R and p.W237C, respectively, at the same position of anosmin-1 protein. The two novel variants had not been presented in the normal population by searching databases (1000 Genomes, EXAC, ESP6500, gnomAD, and dbSNP) (Table S2), and the W239 of anosmin-1 is highly conserved in multiple species (Fig. S1). Both p.W237R and p.W237C were predicted to be damaging using multiple software programs, such as Revel, SIFT, PolyPhen-2, MutationTaster, M-CAP, GERP, and PROVEAN (Table S2).
Pathogenicity analysis of FRFR1 mutations
FRFR1 c.1835delA variation in family 4 was a frameshift mutation that resulted in conversion of glutamate-612 to glycine and generated a stop codon at amino acid 631 (p.Glu612Glyfs*20). According to the ACMG guideline, FRFR1 c.1835delA was assessed as a pathogenicity mutation using the evidence of PVS1 (null variant), PS2 (de novo variant), and PM2 (absent from controls). FRFR1 p.Glu612Glyfs*20 mutation could express a truncated protein lacking the second tyrosine kinase domain (TKD) (Fig. 5B).
Chromosome 8p12-p11.22 gene map and the topological structure of FGFR1. A The orange box in the chromosome 8 diagram at the top indicated the region highlighted below. The OMIM morbid genes are listed at the bottom. B Membrane topology structure of full length FGFR1. Each circle represented a residue with the one-letter symbol. The different domains were labeled as indicated. Locations of pathogenic variations were colored as indicated. and the red box marked the amino acids that affected by FRFR1 c.1835delA mutation
The CNV of seq[GRCh37]del(8)(p12p11.22)chr8:g.36140000_38460000del in patient 5 contained 19 protein-coding genes, including 6 morbid genes of the Online Mendelian Inheritance in Man (OMIM) database (Fig. 5A). The CNV was judged as a pathogenic variation using the evidence of 2A (complete overlap of an established haploinsufficiency gene FGFR1 which contributes to KS2 [ClinGen ID: ISCA-32176], 1.00 point) according to the ACMG & ClinGen technical standards.
Discussion
IHH is an inheritable disorder with clinical and genetic heterogeneity. The major pathogenesis of IHH is failure to activate pulsatile secretion of gonadotropin-releasing hormone (GnRH) during puberty [14]. As the main category of IHH, KS exhibits the typical characteristics of gonadal dysplasia and anosmia [15]. Complex symptoms hinder clinicians from making an accurate diagnosis of KS, while ultrasonography, magnetic resonance imaging, and serum hormone level measurements are routine auxiliary diagnostic methods. To date, more than 20 pathogenic genes have been found to be associated with KS, including ANOS1, FGFR1, PROK2, PROKR2, NELF, KISSR1, CHD7, SEMA3A, and FGF8 [16]. Genetic analysis is the core diagnostic method of KS.
Anosmin-1 encoded by ANOS1 plays important roles in substrate adhesion and cell migration of GnRH-1 neurons, axon outgrowth, and collateral formation, which is basically related to the central nervous system [17,18,19]. Herein, we reported three novel ANOS1 mutations. One of them, ANOS1 c.1063-2 A > T mutation in family 1 led to the production of a new transcript with c.1063–1086 deletion by the transcriptional splicing verification, and a truncated protein with p.355–362 deletion was predicted. It has been known that mutations that affect the splice donor and acceptor site (canonical GT-AG) are highly predictive of splicing defects, and always lead to an abnormal transcript with complete skipping of the downstream exon [20]. However, it is also possible that some splice acceptor site mutations disrupt the original splice acceptor site, and activate a cryptic splice acceptor site, resulting in an aberrant splicing with partial deletion of the downstream exon [21]. Therefore, the verification of a novel splice site variant is essential to clarify the pathogenic mechanism. In the present study, ANOS1 c.1063-2 A > T mutation broken the splice acceptor site AG at intron 7, substituted by TG, and activated a cryptic splice acceptor site AG in exon 8. Intriguingly, both normal and aberrant transcripts of ANOS1 were detected in the proband’s peripheral blood. Previous studies have reported that males carrying a hemizygous splicing mutation of genes located on chromosome X showed both normal and aberrant transcripts, such as c.5786 + 4 A > G in ATRX, c.1044 + 5 G > A in FOXP3, and c.463-6 T > G in OGT [22,23,24], so far the underlying mechanism remained unclear. We speculated that the introduced non-canonical splice site GT-TG caused by the c.1063-2 A > T mutation could produce the normal transcript as GT-TG splice site had been found in animal genomes [25]. Moreover, skewed X-chromosome inactivation (XCI) may be the reason of the few aberrant transcripts in the maternal peripheral blood. Polla et al. reported that the aberrant transcript of MED12 in the peripheral blood samples of those heterozygous women could not be detected because of the skewed XCI [26].
Additionally, we reported two novel missense mutations of p.W237C in family 2 and p.W237R in family 3, which changed the amino acid sequence at the same position of anosmin-1 protein to different amino acid substitutions. W237 was located within the FnIII.1 domain with a highly conservatism. Anosmin-1 is a component of the FGFR1 pathway [27]. WAP, FnIII.1 FnIII.3, and FnIII.4 domains of anosmin-1 were confirmed to interact with FGFR1 [28, 29], and relevant mutations such as N267K in FnIII.1, E514K and F517L in FnIII.3 were proved to influence their interaction [30]. p.W237C and p.W237R may also regulate the signal transduction of the FGFR1 pathway and lead to disease occurrence. Notably, the mutations carried by patients in families 2 and 3 led to different clinical manifestations. Both patients in family 3 showed unilateral deletion of the left kidney, while patients in family 2 had healthy kidney, which attracted our attention to study the relationship between anosmin-1 and kidney development. Approximately 10% of males with ANOS1 pathogenic variants also showed unilateral renal agenesis [31]. Georgopoulos et al. proposed that the FnIII repeats of ANOS1 might be essential for normal renal development [32], but the specific mechanism has not been fully elucidated. Our findings may provide a new idea for the mechanism of anosmin-1 involvement in kidney development.
FGFR1 is responsible for an autosomal dominant form of KS. A full-length FGFR1 protein consists of an extracellular region, composed of three immunoglobulin-like domains (IgI, IgII, and IgIII) responsible for the receptor’s affinity and specificity to its ligands, a single hydrophobic membrane-spanning segment, and two cytoplasmic TKDs with tyrosine kinase activity (Fig. 5B) [33]. The analysis of the total 198 FGFR1 missense mutations listed in the HGMD database showed no obvious characteristics. The overall mutation locations are relatively scattered, mainly in the IgG-like domains and intracellular TKDs, but rarely in the transmembrane region (Fig. 5B). Herein, we identified a novel frameshift FGFR1 mutation c.1835delA (p.Glu612Glyfs*20) in family 4. Following stop codons mutations of FGFR1 including p.R661* and p.Q680* were considered as pathogenic mutations, with protein truncation and nonsense-mediated mRNA decay (NMD) being the underlying mechanisms [34, 35]. Similarly, FGFR1 p.Glu612Glyfs*20 mutation was supposed to result in the synthesis of a truncated inactive receptor lacking an essential portion of the catalytic TKD. Alternatively, the FGFR1 mRNA containing the premature stop codon could be degraded through NMD, leading to haploinsufficiency. In addition, we reported a family (family 5) with a CNV covering the entire FGFR1. To our knowledge, the whole FGFR1 gene deletion was previously reported in only seven cases of KS [9, 33, 36, 37]. By searching the ClinVar database, only six cases with complete FGFR1 were included (VCV000147723, VCV000057114, VCV000150494, VCV000145616, VCV000060360, VCV000362858, VCV000282690, and VCV000395746). The ClinGen database revealed that the haploinsufficiency of FGFR1 contributed to KS2. The 2.32 Mb deletion at 8p12-p11.22 (36,140,000–38,460,000) in the proband of family 5 covered the whole FGFR1 gene. This rare CNV was judged as the pathogenic mutation of KS in this patient as a sporadic case, which is the first report of whole FGFR1 deletion in Chinese population.
Conclusions
In this study, we identified three novel pathogenic mutations of ANOS1 and two novel mutations of FGFR1 in five Chinese families. In silico prediction indicated that ANOS1 c.711 G > T and c.709 T > A were pathogenic mutations. Splicing experiments elucidated the pathogenesis of the ANOS1 c.1063-2 A > T mutation. FRFR1 c.1835delA mutation and del(8)(p12p11.22)chr8:g.36140000_38460000del were assessed as pathogenic variations. Therefore, our study expands the spectrum of mutations associated with KS and provides diagnostic evidence for patients who carry the same mutation in the future.
Availability of data and materials
The data analyzed during the current study is available from the corresponding author on reasonable request.
Abbreviations
- KS:
-
Kallmann syndrome
- IHH:
-
Idiopathic hypogonadotropic hypogonadism
- NGS:
-
Next-generation sequencing
- HGMD:
-
Human Gene Mutation Database
- FGFR1:
-
Fibroblast growth factor receptor 1
- DSD:
-
Disorder of sex development
- GnRH:
-
Gonadotropin-releasing hormone
- XCI:
-
X-chromosome inactivation
- TKD:
-
Tyrosine kinase domain
- NMD:
-
Nonsense-mediated mRNA decay
References
Quinton R, Duke VM, Robertson A, Kirk JM, Matfin G, de Zoysa PA, et al. Idiopathic gonadotrophin deficiency: genetic questions addressed through phenotypic characterization. Clin Endocrinol (Oxf). 2001;55:163–74. https://doi.org/10.1046/j.1365-2265.2001.01277.x.
Boehm U, Bouloux PM, Dattani MT, de Roux N, Dodé C, Dunkel L, et al. Expert consensus document: European Consensus Statement on congenital hypogonadotropic hypogonadism–pathogenesis, diagnosis and treatment. Nat Rev Endocrinol. 2015;11:547–64. https://doi.org/10.1038/nrendo.2015.112.
Laitinen EM, Vaaralahti K, Tommiska J, Eklund E, Tervaniemi M, Valanne L, et al. Incidence, phenotypic features and molecular genetics of Kallmann syndrome in Finland. Orphanet J Rare Dis. 2011;6:41. https://doi.org/10.1186/1750-1172-6-41.
Stamou MI, Georgopoulos NA. Kallmann syndrome: phenotype and genotype of hypogonadotropic hypogonadism. Metabolism. 2018;86:124–34. https://doi.org/10.1016/j.metabol.2017.10.012.
Jiang X, Li D, Gao Y, Zhang X, Wang X, Yang Y, et al. A novel splice site variant in ANOS1 gene leads to Kallmann syndrome in three siblings. Gene. 2020;726:144177. https://doi.org/10.1016/j.gene.2019.144177.
Franco B, Guioli S, Pragliola A, Incerti B, Bardoni B, Tonlorenzi R, et al. A gene deleted in Kallmann’s syndrome shares homology with neural cell adhesion and axonal path-finding molecules. Nature. 1991;353:529–36. https://doi.org/10.1038/353529a0.
del Castillo I, Cohen-Salmon M, Blanchard S, Lutfalla G, Petit C. Structure of the X-linked Kallmann syndrome gene and its homologous pseudogene on the Y chromosome. Nat Genet. 1992;2:305–10. https://doi.org/10.1038/ng1292-305.
Nie M, Xu H, Chen R, Mao J, Wang X, Xiong S, et al. Analysis of genetic and clinical characteristics of a Chinese Kallmann syndrome cohort with ANOS1 mutations. Eur J Endocrinol. 2017;177:389–98. https://doi.org/10.1530/EJE-17-0335.
Dodé C, Levilliers J, Dupont JM, De Paepe A, Le Dû N, Soussi-Yanicostas N, et al. Loss-of-function mutations in FGFR1 cause autosomal dominant Kallmann syndrome. Nat Genet. 2003;33:463–5. https://doi.org/10.1038/ng1122.
Böttcher RT, Niehrs C. Fibroblast growth factor signaling during early vertebrate development. Endocr Rev. 2005;26:63–77. https://doi.org/10.1210/er.2003-0040.
Gach A, Pinkier I, Szarras-Czapnik M, Sakowicz A, Jakubowski L. Expanding the mutational spectrum of monogenic hypogonadotropic hypogonadism: novel mutations in ANOS1 and FGFR1 genes. Reprod Biol Endocrinol. 2020;18:8. https://doi.org/10.1186/s12958-020-0568-6.
Zhang R, Chen Z, Song Q, Wang S, Liu Z, Zhao X, et al. Identification of seven exonic variants in the SLC4A1, ATP6V1B1, and ATP6V0A4 genes that alter RNA splicing by minigene assay. Hum Mutat. 2021;42:1153–64. https://doi.org/10.1002/humu.24246.
Okonechnikov K, Golosova O, Fursov M; UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics. 2012;28:1166–7. https://doi.org/10.1093/bioinformatics/bts091.
Bianco SD, Kaiser UB. The genetic and molecular basis of idiopathic hypogonadotropic hypogonadism. Nat Rev Endocrinol. 2009;5:569–76. https://doi.org/10.1038/nrendo.2009.177.
Sarfati J, Bouvattier C, Bry-Gauillard H, Cartes A, Bouligand J, Young J. Kallmann syndrome with FGFR1 and KAL1 mutations detected during fetal life. Orphanet J Rare Dis. 2015;10:71. https://doi.org/10.1186/s13023-015-0287-9.
Chen K, Wang H, Lai Y. Kallmann Syndrome Due to Heterozygous Mutation in SOX10 Coexisting With Waardenburg Syndrome Type II: Case Report and Review of Literature. Front Endocrinol (Lausanne). 2021;11:592831. https://doi.org/10.3389/fendo.2020.592831.
Hamada AJ, Esteves SC, Agarwal A. A comprehensive review of genetics and genetic testing in azoospermia. Clinics (Sao Paulo). 2013;68:39–60. https://doi.org/10.6061/clinics/2013(sup01)06.
Cariboni A, Pimpinelli F, Colamarino S, Zaninetti R, Piccolella M, Rumio C, et al. The product of X-linked Kallmann’s syndrome gene (KAL1) affects the migratory activity of gonadotropin-releasing hormone (GnRH)-producing neurons. Hum Mol Genet. 2004;13:2781–91. https://doi.org/10.1093/hmg/ddh309.
Soussi-Yanicostas N, Faivre-Sarrailh C, Hardelin JP, Levilliers J, Rougon G, Petit C. Anosmin-1 underlying the X chromosome-linked Kallmann syndrome is an adhesion molecule that can modulate neurite growth in a cell-type specific manner. J Cell Sci. 1998;111:2953–65.
Saadah OI, Banaganapalli B, Kamal NM, Sahly AN, Alsufyani HA, Mohammed A, et al. Identification of a Rare Exon 19 Skipping Mutation in ALMS1 Gene in Alström Syndrome Patients From Two Unrelated Saudi Families. Front Pediatr. 2021;9:652011. https://doi.org/10.3389/fped.2021.652011.
Thongnoppakhun A, Rungroj N, Wilairat P, Vareesangthip K, Sirinavin C, Yenchitsomanus PT. A novel splice-acceptor site mutation (IVS13-2A>T) of polycystic kidney disease 1 (PKD1) gene resulting in an RNA processing defect with a 74-nucleotide deletion in exon 14 of the mRNA transcript. Hum Mutat. 2000;15:115. https://doi.org/10.1002/(SICI)1098-1004(200001)15:1%3c115::AID-HUMU22%3e3.0.CO;2-Z.
Cong Y, Wu J, Wang H, Wu K, Huang C, Yang X. Identification of a Hemizygous Novel Splicing Variant in ATRX Gene: A Case Report and Literature Review. Front Pediatr. 2022;10:834087. https://doi.org/10.3389/fped.2022.834087.
Hwang JL, Park SY, Ye H, Sanyoura M, Pastore AN, Carmody D, et al. FOXP3 mutations causing early-onset insulin-requiring diabetes but without other features of immune dysregulation, polyendocrinopathy, enteropathy. X-linked syndrome Pediatr Diabetes. 2018;19:388–92. https://doi.org/10.1111/pedi.12612.
Willems AP, Gundogdu M, Kempers MJE, Giltay JC, Pfundt R, Elferink M, et al. Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J Biol Chem. 2017;292:12621–31. https://doi.org/10.1074/jbc.M117.790097.
Frey K, Pucker B. Animal, Fungi, and Plant Genome Sequences Harbor Different Non-Canonical Splice Sites. Cells. 2020;9:458. https://doi.org/10.3390/cells9020458.
Polla DL, Bhoj EJ, Verheij JBGM, Wassink-Ruiter JSK, Reis A, Deshpande C, et al. De novo variants in MED12 cause X-linked syndromic neurodevelopmental disorders in 18 females. Genet Med. 2021;23:645–52. https://doi.org/10.1038/s41436-020-01040-6.
Ayari B, Soussi-Yanicostas N. FGFR1 and anosmin-1 underlying genetically distinct forms of Kallmann syndrome are co-expressed and interact in olfactory bulbs. Dev Genes Evol. 2007;217:169–75. https://doi.org/10.1007/s00427-006-0125-0.
Murcia-Belmonte V, Astillero-López V, Esteban PF. Anosmin 1 Interacts with the Prokineticin Receptor 2 In Vitro Indicating a Molecular Link Between Both Proteins in the Pathogenesis of Kallmann Syndrome. Protein Pept Lett. 2016;23:650–5. https://doi.org/10.2174/0929866523666160517123331.
Murcia-Belmonte V, Esteban PF, García-González D, De Castro F. Biochemical dissection of Anosmin-1 interaction with FGFR1 and components of the extracellular matrix. J Neurochem. 2010;115:1256–65. https://doi.org/10.1111/j.1471-4159.2010.07024.x.
de Castro F, Esteban PF, Bribián A, Murcia-Belmonte V, García-González D, Clemente D. The adhesion molecule anosmin-1 in neurology: Kallmann syndrome and beyond. Adv Neurobiol. 2014;8:273–92. https://doi.org/10.1007/978-1-4614-8090-7_12.
Louden ED, Poch A, Kim HG, Ben-Mahmoud A, Kim SH, Layman LC. Genetics of hypogonadotropic Hypogonadism-Human and mouse genes, inheritance, oligogenicity, and genetic counseling. Mol Cell Endocrinol. 2021;534:111334. https://doi.org/10.1016/j.mce.2021.111334.
Georgopoulos NA, Koika V, Galli-Tsinopoulou A, Spiliotis BE, Adonakis G, Keramida MK, et al. Renal dysgenesis and KAL1 gene defects in patients with sporadic Kallmann syndrome. Fertil Steril. 2007;88:1311–7. https://doi.org/10.1016/j.fertnstert.2006.12.044.
Sato N, Katsumata N, Kagami M, Hasegawa T, Hori N, Kawakita S, et al. Clinical assessment and mutation analysis of Kallmann syndrome 1 (KAL1) and fibroblast growth factor receptor 1 (FGFR1, or KAL2) in five families and 18 sporadic patients. J Clin Endocrinol Metab. 2004;89:1079–88. https://doi.org/10.1210/jc.2003-030476.
Dodé C, Fouveaut C, Mortier G, Janssens S, Bertherat J, Mahoudeau J, et al. Novel FGFR1 sequence variants in Kallmann syndrome, and genetic evidence that the FGFR1c isoform is required in olfactory bulb and palate morphogenesis. Hum Mutat. 2007;28:97–8. https://doi.org/10.1002/humu.9470.
Pitteloud N, Acierno JS Jr, Meysing A, Eliseenkova AV, Ma J, Ibrahimi OA, et al. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A. 2006;103:6281–6. https://doi.org/10.1073/pnas.0600962103.
Wang D, Lai P. Global retardation and hereditary spherocytosis associated with a novel deletion of chromosome 8p11.21 encompassing KAT6A and ANK1. Eur J Med Genet. 2020;63:104082. https://doi.org/10.1016/j.ejmg.2020.104082.
Trarbach EB, Teles MG, Costa EM, Abreu AP, Garmes HM, Guerra G Jr, et al. Screening of autosomal gene deletions in patients with hypogonadotropic hypogonadism using multiplex ligation-dependent probe amplification: detection of a hemizygosis for the fibroblast growth factor receptor 1. Clin Endocrinol (Oxf). 2010;72:371–6. https://doi.org/10.1111/j.1365-2265.2009.03642.x.
Acknowledgements
The authors would like to thank all the families for their cooperation in this study and Professor Leping Shao at The Affiliated Qingdao Municipal Hospital of Qingdao University for kindly providing the pSPL3 vector.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 32000451), the National Natural Science Foundation of China (Grant No. 82001497), 345 Talent Project of Shengjing Hospital (M1335), and 345 Talent Project of Shengjing Hospital (M1328).
Author information
Authors and Affiliations
Contributions
Y. Z., P. L., and G. C. conceived and designed this study. Q. Z. and R. H. recruited the patients and collected peripheral blood samples. G. C. and P. L. performed the experiments. P. L. and Y. Z. analyzed the data. Y. Z. supervised the research. G. C. wrote the original draft. P. L. edited the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the Medical Ethics Committee of Shengjing Hospital of China Medical University (approval no. 2021PS548K), and informed consent was obtained from the families.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1:
Fig. S1 Conservation analysis of anosmin-1. Assessment of amino acid conservation of anosmin-1 using Ugene. W237 of anosmin-1 (indicated in a black box) is highly conserved across various species.
Additional file 2: Table S1
The clinical and genetic data of patients.
Additional file 3: Table S2
The results of population databases retrieval and pathogenicity prediction for ANOS1 gene c.709 T > A and c.711 G > T variation.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chu, G., Li, P., Zhao, Q. et al. Mutation spectrum of Kallmann syndrome: identification of five novel mutations across ANOS1 and FGFR1. Reprod Biol Endocrinol 21, 23 (2023). https://doi.org/10.1186/s12958-023-01074-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12958-023-01074-w